I want to crop a raster using a bbox or a known extent, i.e., 10 pixels in row and col.
Below you can see a reproducible example:
library(terra)
r <- terra::set.ext(rast(volcano), terra::ext(0, nrow(volcano), 0, ncol(volcano)))
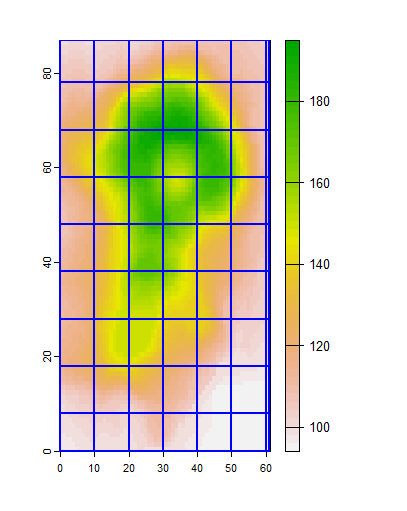
plot(r)
xmin <- xmin(r)
ymax <- ymax(r)
rs <- res(r)
n <- 10 # i.e. of rows and cols size
xmax <- xmin n * rs[1]
ymin <- ymax - n * rs[2]
x <- crop(r, c(xmin, xmax, ymin, ymax))
plot(x)
The intended loop is to:
- To go through all the raster (
r) length cropping and save each raster piece temporarily into data.frame (or data.table, raster, spatraster, list)
CodePudding user response:
It would be useful to have more context about why you would do this. It is also not clear from your question if you want the cropped areas to overlap; I assume you do not want that.
You could so something like:
library(terra)
r <- rast(volcano)
n <- 10
Get the starting cells of interest
rows <- seq(1, nrow(r), by=n)
cols <- seq(1, ncol(r), by=n)
cells <- cellFromRowColCombine(r, rows, cols)
Get the coordinates
# upper-left coordinates of the starting cells
xy <- xyFromCell(r, cells)
rs <- res(r)
xy[,1] <- xy[,1] - rs[1]/2
xy[,2] <- xy[,2] rs[2]/2
# add the lower-right coordinates of the end cell
xy <- cbind(xy[,1], xy[,1] n*rs[1], xy[,2] - n*rs[2], xy[,2])
And loop
x <- lapply(1:nrow(xy), function(i) {
crop(r, xy[i,])
})
Verify
e <- lapply(x, \(i) ext(i) |> as.polygons()) |> vect()
plot(r)
lines(e, col="blue", lwd=2)
sapply(x, dim) |> t() |> head()
# [,1] [,2] [,3]
#[1,] 10 10 1
#[2,] 10 10 1
#[3,] 10 10 1
#[4,] 10 10 1
#[5,] 10 10 1
#[6,] 10 10 1
Or use an alternative approach based on the start- and end-cell numbers
srows <- seq(1, nrow(r), by=n)
scols <- seq(1, ncol(r), by=n)
erows <- pmin(nrow(r), rows n-1)
ecols <- pmin(ncol(r), cols n-1)
scell <- cellFromRowColCombine(r, rows, cols)
ecell <- cellFromRowColCombine(r, erows, ecols)
cells <- cbind(scell, ecell)
x <- lapply(1:nrow(cells), function(i) {
e <- ext(r, cells[i,])
crop(r, e)
})
CodePudding user response:
Using terra data, by your approach, which I like:
library(terra)
r <- terra::set.ext(rast(volcano), terra::ext(0, nrow(volcano), 0, ncol(volcano)))
Then we need extents that have 4 values (xmin,xmax,ymin,ymax), [for indices {i,j,k,l}} we then want to traverse the raster from top left to top right, then again stepping down, and down till we get to bottom right [for indices {o,p}], then a further crop [for index {q}]. This is a pretty deep nesting for for loops, and in the interest of preserving mental...it might be better to make some helpers.
x_mtx <- matrix(c(seq(0,70,10), seq(10,80,10)), ncol = 2)
y_mtx <- matrix(c(seq(51, 1, -10), seq(61, 11, -10)), ncol = 2)
these eliminate indices i:l, then we make our extents:
crop_exts <- vector(mode = 'list', length= 48L)
for(p in 1:6){
for(o in 1:8){
crop_exts[[p]][[o]] <- ext(x_mtx[o, 1], x_mtx[o, 2], y_mtx[p, 1], y_mtx[p, 2])
}
}
# check our work
crop_exts[[1]][[1]] |> as.vector()
#xmin xmax ymin ymax
#0 10 51 61
crop_exts[[6]][[8]] |> as.vector()
#xmin xmax ymin ymax
# 70 80 1 11
Looks like top left to bottom right. And noting the [[1]][[1]], [[6]][[8]], we have a second nested {p, o} loop as above in view of our indexing here to pull out the crops:
for(p in 1:6){
for(o in 1:8){
cropped_r[[p]][[o]] <- crop(r, crop_exts[[p]][[o]])
}
}
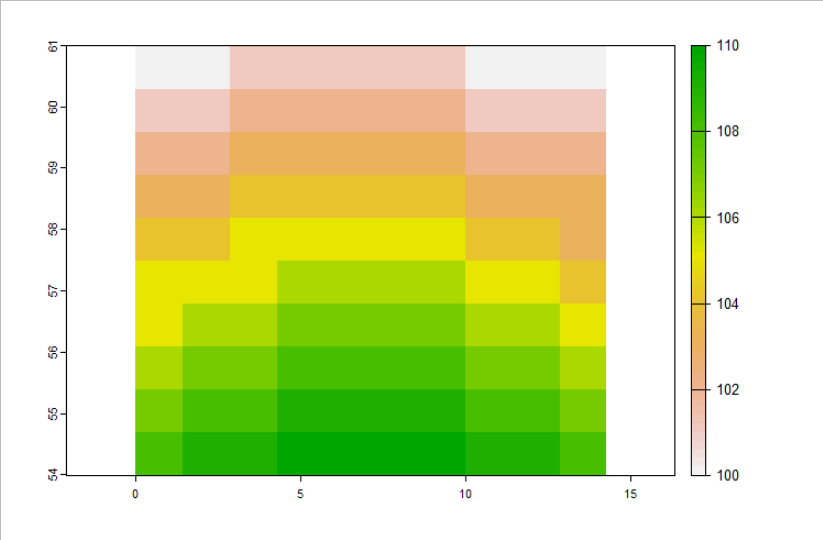
plot(cropped_r[[1]][[1]])
plot(cropped_r[[6]][[8]])
Possibly easier, overall, is to generate the complete extent matrix, dim(48,4) in this case, but couldn't wrap my head around it, and keep in mind indices, and preserve my mental...