I'm having trouble working with missing data in my csv. I would like for the output below to look the same, just without the nan labels, but all preserve the state borders with the missing data.
Output with "nan" and decimal issue:
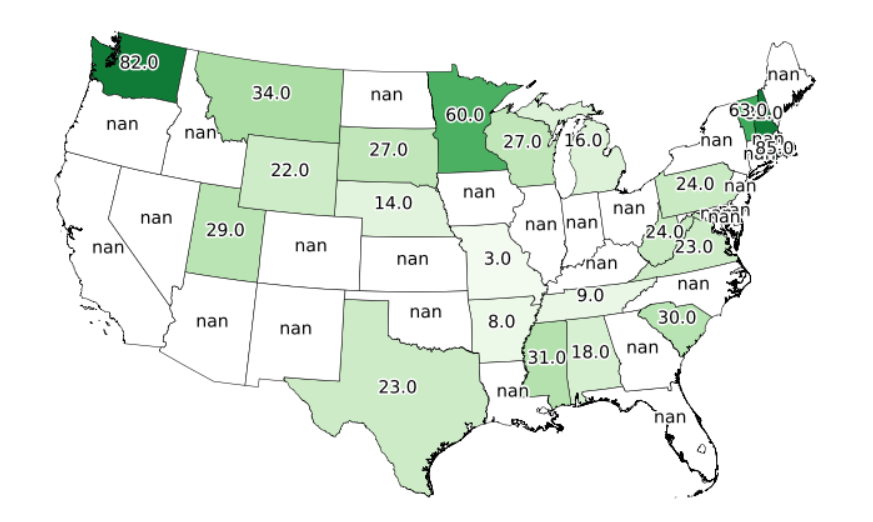
In addition, I'm having trouble with the decimal places. I do not want these to appear. Also, I'm only trying to do this for the specific column of data I am trying to plot. Below is the code I have tried to address both of these issues:
csv = pd.read_csv(r'C:\Downloads\Data.csv')
sf = r'C:\Downloads\s_11au16\s_11au16.shp'
US = gpd.read_file(sf)
#Merge them
data = gpd.GeoDataFrame(csv.merge(US))
#set projection
data = data.to_crs(epsg=6923)
#data = data[['NAME', 'soil_data']]
#data = data[data['soil_data'].notna()]
#data.soil_data = data.soil_data.astype(int)
#set up basemap
ax = data.plot(figsize = (12,8), column="soil_data", cmap="Greens", edgecolor='black', linewidth=.5, vmin=0, vmax=100,missing_kwds={"color": "white", "edgecolor": "k", "label": "none"})
#ax.set_title("Topsoil Moisture: Adequate Surplus %", fontsize=18, fontweight='bold')
ax.set_axis_off()
#annotate data
label = data
label.apply(lambda x: ax.annotate(text=x['soil_data'], xy=x.geometry.centroid.coords[0], color="black", ha='center', fontsize=14,
path_effects=[pe.withStroke(linewidth=3, foreground="white")]), axis=1)
I tried to use this block code below to solve the problem, but this did not work.
data = data[['NAME', 'soil_data']]
data = data[data['soil_data'].notna()]
data.soil_data = data.soil_data.astype(int)
Again, the two problems are 1) getting nan to not label, and 2) make labels whole numbers.
CodePudding user response:
Difficult to be sure without a fully reproducible example, but I would bet for something like:
#annotate data
label = data.dropna(subset='soil_data')
label.apply(lambda x: ax.annotate(text=int(x['soil_data']), xy=x.geometry.centroid.coords[0], color="black", ha='center', fontsize=14,
path_effects=[pe.withStroke(linewidth=3, foreground="white")]), axis=1)
