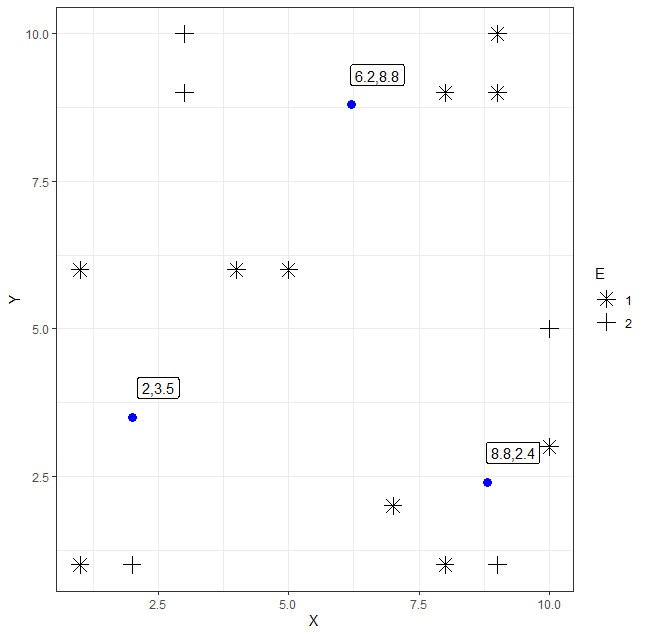
I have some data for which I have performed manual K-means clustering using Euclidean distance. I have the final centroids and the clustered points. I have plotted this to a graph however, I am unsure how I could go about colour coding the different points depending on the cluster they belong to.
This is how the data has been clustered and I would like to colour the points depending on the colour they are in.
Cluster1 centroid location: 2, 3.5
points under Cluster1: (1,1), (1,6), (2,1), (4,6)
Centroid2 centroid location: 6.2, 8.8
points under Cluster2: (3,9), (3,10), (5,6), (8,9), (9,9), (9,10)
Centroid3 centroid location: 8.8, 2.4
points under Cluster3: (7,2), (8,1), (9,1), (10,3), (10,5)
Data
structure(list(Subject = 1:15, X = c(1L, 1L, 2L, 3L, 3L, 4L,
5L, 7L, 8L, 8L, 9L, 9L, 9L, 10L, 10L), Y = c(1L, 6L, 1L, 9L,
10L, 6L, 6L, 2L, 1L, 9L, 1L, 9L, 10L, 3L, 5L), E1 = c(1L, 1L,
NA, NA, NA, 1L, 1L, 1L, 1L, 1L, NA, 1L, 1L, 1L, NA), E2 = c(NA,
NA, 2L, 2L, 2L, NA, NA, NA, NA, NA, 2L, NA, NA, NA, 2L)), class = "data.frame", row.names = c(NA,
-15L))
Current implementation for the graph
library(tidyverse)
library(ggplot2)
data <- read.csv("data.csv")
data2 <- data[, -c(1)]
data2 <- data %>% filter(!if_all(c(E1, E2), is.na)) %>% mutate(E = ifelse(is.na(E1), E2, E1))
# Format centroid positions to be a 2D point with coordinates
x <- c(2, 6.2, 8.8)
y <- c(3.5, 8.8, 2.4)
coords = paste(x,y, sep=",")
df = data.frame(x,y)
ggplot(data2, aes(X, Y, shape = factor(E)))
geom_point(size = 4)
scale_shape_manual(values = c(8, 3), name = "E")
theme_bw()
geom_point(df, mapping = aes(x, y), col = "blue", size = 3, inherit.aes = FALSE)
geom_label(df, mapping = aes(x .5, y 0.5, label = coords), inherit.aes = FALSE)
Current produced graph
CodePudding user response:
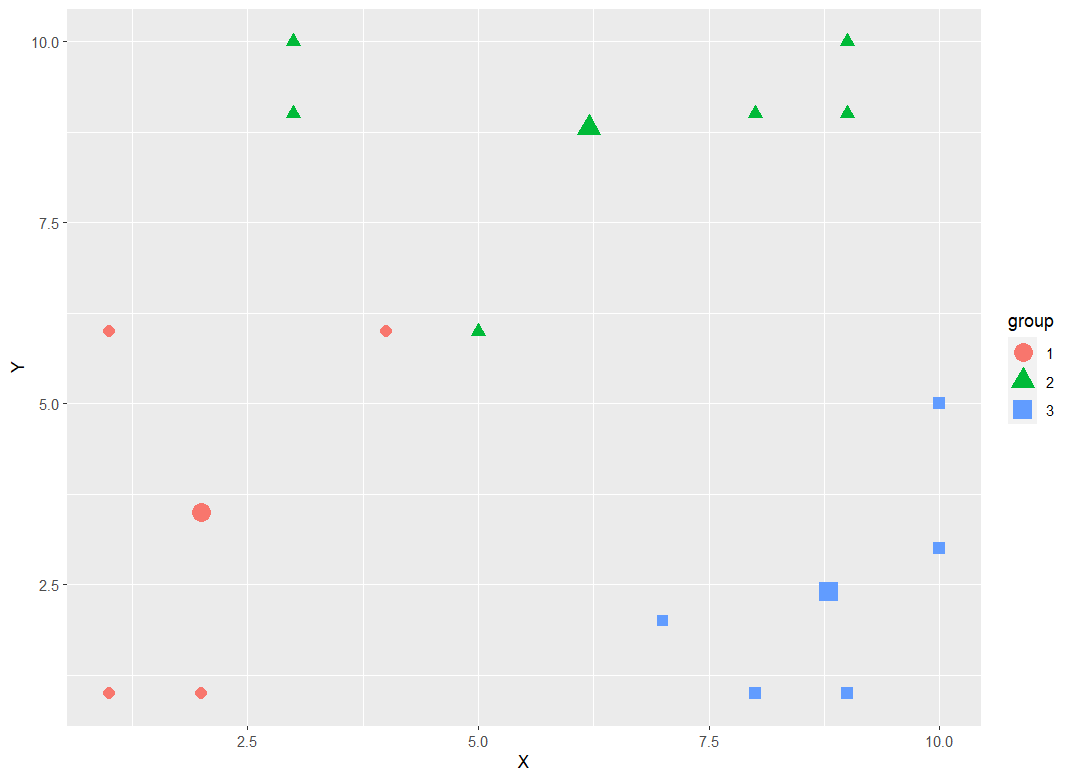
You will need to determine for each point which cluster they belong to and add this information to the plotting dataframe. Below is one approach:
df %>%
# add group
mutate(group = factor(row_number())) %>%
# create all combinations with data2 (rows for each point in data2 with each centroid)
crossing(data2) %>%
# compute euclidean distance
mutate(dist = (X-x)^2 (Y-y)^2) %>%
# for each subject, filter for the closest centroid
group_by(Subject) %>%
slice_min(dist) %>%
ungroup() %>%
# plot
ggplot(aes(colour = group, shape = group))
geom_point(aes(X, Y), size = 3)
geom_point(aes(x, y), size = 5)
CodePudding user response:
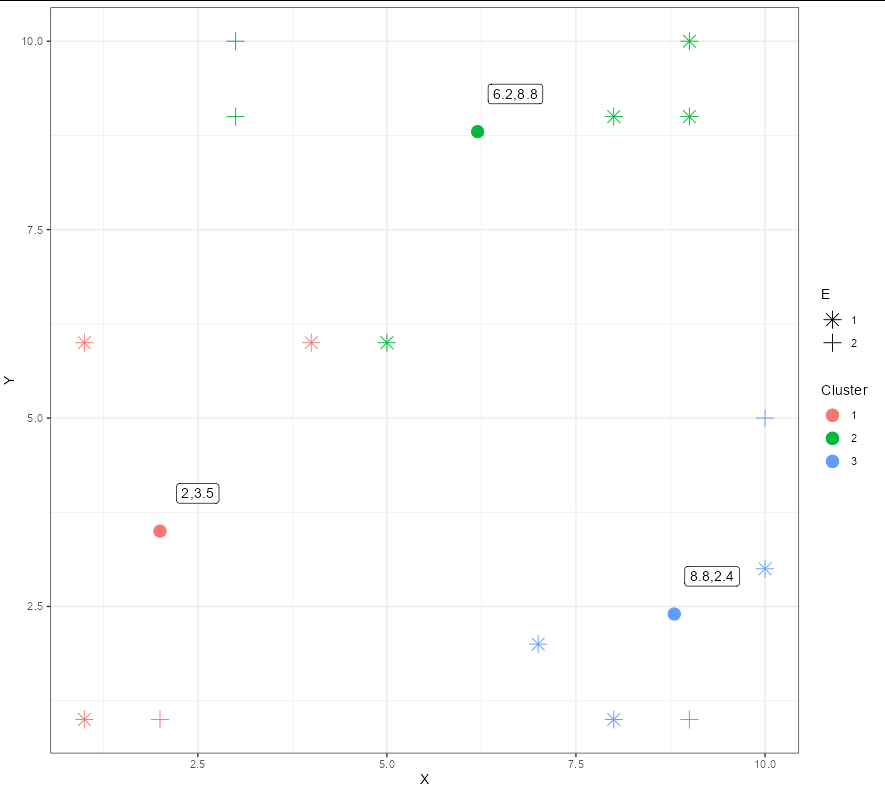
You need to add the cluster information:
data2$Cluster <- factor(c(1, 1, 1, 2, 2, 1, 2, 3, 3, 2, 3, 2, 2, 3, 3))
df$Cluster <- factor(1:3)
Now your plot can be done with:
ggplot(data2, aes(X, Y, shape = factor(is.na(E1))))
geom_point(size = 4, aes(color = Cluster))
scale_shape_manual(values = c(8, 3), name = "E", labels = 1:2)
theme_bw()
geom_point(data = df, aes(x, y, color = Cluster), size = 4,
inherit.aes = FALSE)
geom_label(data = df, aes(x .5, y 0.5, label = coords),
inherit.aes = FALSE)