Hello I have a tibble through a pipe from tidytext::unnest_tokens() and count(category, word, name = "count"). It looks like this example.
owl <- tibble(category = c(0, 1, 2, -1, 0, 1, 2),
word = c(rep("hello", 3), rep("world", 4)),
count = sample(1:100, 7))
and I would like to get this tibble with an additional column that gives the number of categories the word appears in, i.e. the same number for each time the word appears.
I tried the following code that works in principal. The result is what I want.
owl %>% mutate(sum_t = sapply(1:nrow(.), function(x) {filter(., word == .$word[[x]]) %>% nrow()}))
However, seeing that my data has 10s of thousands of rows this takes a rather long time. Is there a more efficient way to achieve this?
CodePudding user response:
We could use add_count:
library(dplyr)
owl %>%
add_count(word)
output:
category word count n
<dbl> <chr> <int> <int>
1 0 hello 98 3
2 1 hello 30 3
3 2 hello 37 3
4 -1 world 22 4
5 0 world 80 4
6 1 world 18 4
7 2 world 19 4
CodePudding user response:
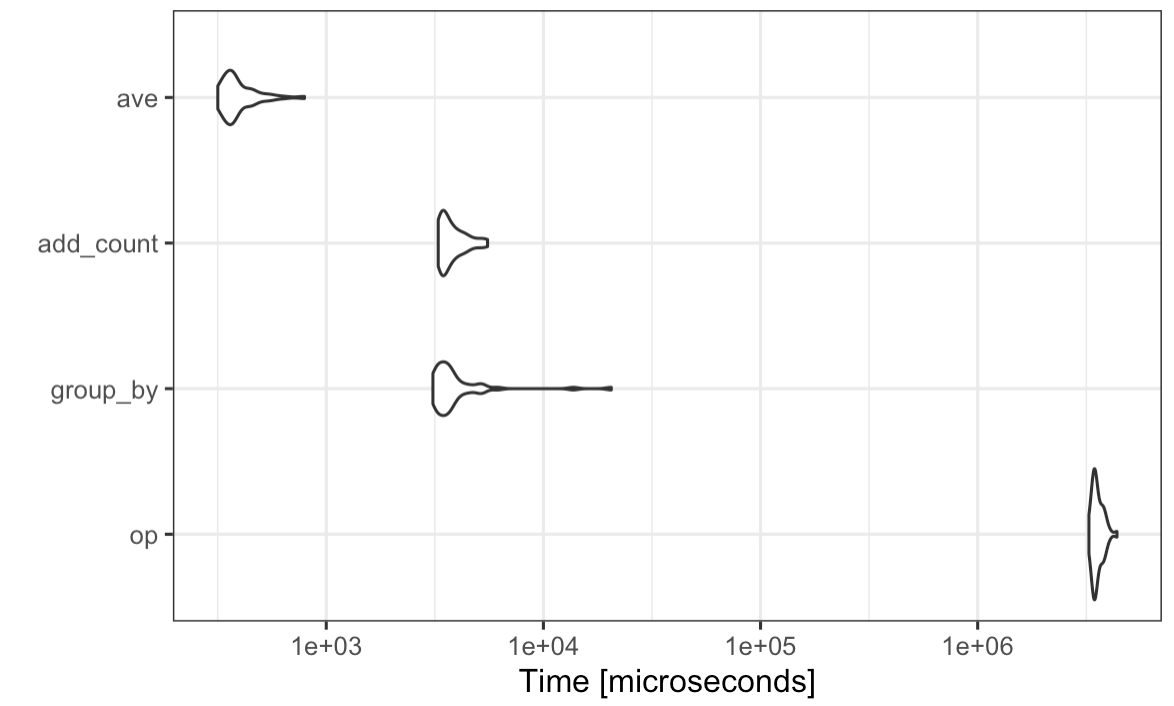
I played around with a few solutions and microbenchmark. I added TarJae's proposition to the benchmark. I also wanted to use the fantastic ave function just to see how it would compare to a dplyr solution.
library(microbenchmark)
n <- 500
owl2 <- tibble(
category = sample(-10:10, n , replace = TRUE),
word = sample(stringi::stri_rand_strings(5, 10), n, replace = TRUE),
count = sample(1:100, n, replace = TRUE))
mb <- microbenchmark(
op = owl2 %>% mutate(sum_t = sapply(1:nrow(.), function(x) {filter(., word == .$word[[x]]) %>% nrow()})),
group_by = owl2 %>% group_by(word) %>% mutate(n = n()),
add_count = owl2 %>% add_count(word),
ave = cbind(owl2, n = ave(owl2$word, owl2$word, FUN = length)),
times = 50L)
autoplot(mb) theme_bw()
The conclusion is that the elegant solution using add_count will save you a lot of time, and, ave speeds up a lot the process.