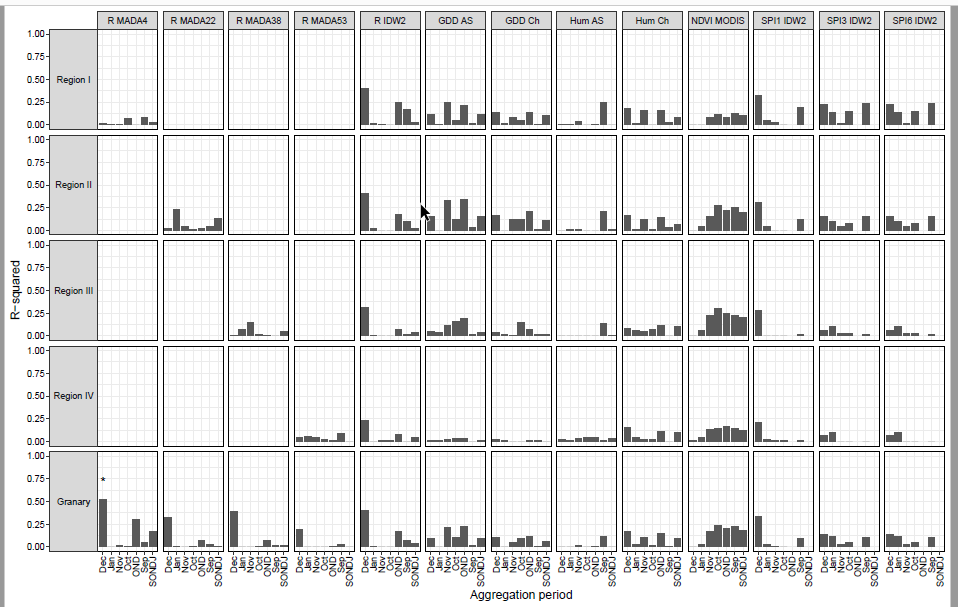
I have a lm model results containing R2 and pvalue, and I plotted them in a bar plot. I have then facetted them using two discrete variables.
I want to put * on the top of bars to flag statistical significance (pvlue <= 0.05), as shown on the bottom-left-most panel of the below image.
I have not found an insightful tutorial on how to do this.
Any way to do this, please?
Here is some code I used
> head(res_all_s2)
WI aggre_per Season yield_level slope Intercept r.squared
1 R IDW2 Dec Season2 Region II -7.06 6091 0.41
2 R IDW2 Dec Season2 Region I -7.29 6280 0.40
3 GDD AS OND Season2 Region II 14.23 -18270 0.34
4 GDD AS Nov Season2 Region II 36.84 -14760 0.33
5 SPI1 IDW2 Dec Season2 Region II -405.10 5358 0.31
6 SPI1 IDW2 Dec Season2 Region I -421.70 5523 0.32
adj.r.squared fstatistic.value pval pearson
1 0.36 9.58 0.01 -0.64
2 0.36 9.49 0.01 -0.64
3 0.29 7.09 0.02 0.58
4 0.28 6.97 0.02 0.58
5 0.26 6.40 0.02 -0.56
6 0.27 6.51 0.02 -0.56
> # significance (pval <= 0.05)
> signif_reg <- res_all_s2 %>% filter(pval <= 0.05)
> head(signif_reg)
WI aggre_per Season yield_level slope Intercept r.squared
1 R IDW2 Dec Season2 Region II -7.06 6091 0.41
2 R IDW2 Dec Season2 Region I -7.29 6280 0.40
3 GDD AS OND Season2 Region II 14.23 -18270 0.34
4 GDD AS Nov Season2 Region II 36.84 -14760 0.33
5 SPI1 IDW2 Dec Season2 Region II -405.10 5358 0.31
6 SPI1 IDW2 Dec Season2 Region I -421.70 5523 0.32
adj.r.squared fstatistic.value pval pearson
1 0.36 9.58 0.01 -0.64
2 0.36 9.49 0.01 -0.64
3 0.29 7.09 0.02 0.58
4 0.28 6.97 0.02 0.58
5 0.26 6.40 0.02 -0.56
6 0.27 6.51 0.02 -0.56
>
> # Plot R2
>
> r <- res_all_s2 %>% ggplot(aes(x=aggre_per,
y=r.squared ))
geom_bar(stat="identity", width=0.8)
facet_grid(yield_level ~ WI,
scales = "free_y",
switch = "y")
scale_y_continuous(limits = c(0, 1))
xlab("Aggregation period")
ylab(expression(paste("R-squared")))
theme_bw()
theme(axis.title = element_text(size = 12), # all titles
axis.text = element_text(colour = "black"),
axis.text.x = element_text(angle = 90, vjust = 0.5,
hjust = 1, color = "black"),
strip.text.y.left = element_text(angle = 0),
panel.border = element_rect(color = "black",
size = .5))
> r
And, here is the link to my res_all_s2 dataset https://1drv.ms/u/s!Ajl_vaNPXhANgckJeqDKA0fzfFEbhg?e=VfoFaB
CodePudding user response:
Technically, you can always add an appropriate geom with its independent dataset (that would be your data filtered to exclude pval > .05):
df_filtered <- res_all_s2 %>% filter(...)
## ggplot(...)
geom_point(data = df_filtered, pch = 8)
## pch = point character, no. 8 = asterisk
or
## ...
geom_text(data = df_filtered, aes(label = '*'), nudge_y = .05)
## nudge_y = vertical offset
or color only significant columns:
## ...
geom_col(aes(fill = c('grey','red')[1 pval <= .05]))
So, yes, technically that's feasible. But before throwing the results of 13 x 7 x 5 = 455 linear models at your audience, please consider the issues of p-hacking, the benefits of multivariate analysis and the viewers' ressources ;-)