I am a compete novice at using loops and simplifying my code.
I have a dataset of places with accompanying data that I would like to plot on separate maps. The individual plotting of the maps I can do.
However I would like to automate the process a little bit, I have new data arriving each day and don't want to repeat the process of cleaning the data and rewriting code.
So I thought a for loop might be the answer
What I need are separate plots for each Time in the data below
So the loop would pull out all the data for each value of Time and then plot it.
dput(df)
structure(list(Site = c("O242", "O51", "O59", "O71", "C110",
"C116", "C120", "C13", "C132", "C134", "C139", "C140", "C29",
"C30", "C33", "C48", "C56", "C9A", "MP25", "MP67", "B30", "MP2",
"B101", "B11", "B112", "B15", "B197", "B2", "B217", "B22", "B30",
"B95", "MP21", "MP25", "MP33", "MP51", "MP56", "MP6", "MP60",
"MP61", "MP67", "MP77", "EX84", "EX92", "SW130", "O31", "O38",
"O38B", "O48", "O58", "O59", "O68", "O71", "O72", "O81", "O94",
"O207", "O209", "O210", "O215"), Time = c(-25, -22, -22, -22,
-14, -14, -14, -14, -14, -14, -14, -14, -14, -14, -14, -14, -14,
-14, -23, -23, -20, -20, -11, -11, -11, -11, -11, -11, -11, -11,
-11, -11, -10, -10, -10, -10, -10, -10, -10, -10, -10, -10, -10,
-10, -10, -10, -10, -10, -10, -10, -10, -10, -10, -10, -10, -10,
-10, -10, -10, -10), Code = c(1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L,
2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 3L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 3L, 2L, 1L, 2L, 1L, 2L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 2L, 1L, 2L, 1L, 3L, 2L, 1L, 3L, 1L, 1L, 3L, 2L, 2L,
3L, 1L, 1L, 2L), lon = c(-1.341280663, -1.343562025, -1.343620358,
-1.340629756, -1.332551665, -1.329108814, -1.328655294, -1.330835311,
-1.330715028, -1.33052464, -1.328144549, -1.328287425, -1.329353862,
-1.329343236, -1.33041446, -1.325353001, -1.327279282, -1.332909331,
-1.300122834, -1.299148682, -1.310197641, -1.305886812, -1.308725397,
-1.309505208, -1.309235075, -1.308580716, -1.30959055, -1.308685087,
-1.309426224, -1.306562029, -1.310197641, -1.307564253, -1.301598673,
-1.300122834, -1.299510666, -1.299846899, -1.297823339, -1.305388627,
-1.297220016, -1.297398331, -1.299148682, -1.300378324, -1.333554619,
-1.338688389, -1.332015649, -1.344951753, -1.344769267, -1.345214102,
-1.342514477, -1.343145083, -1.343620358, -1.34275518, -1.340629756,
-1.339067762, -1.338035147, -1.335442485, -1.346461847, -1.34550727,
-1.34516939, -1.346584124), lat = c(51.76635545, 51.76553293,
51.76450781, 51.76428383, 51.75689245, 51.75615401, 51.75742817,
51.75637019, 51.75666667, 51.75740286, 51.7596281, 51.75976378,
51.75721637, 51.75695556, 51.75701561, 51.75871255, 51.75875955,
51.75720018, 51.76339382, 51.75986347, 51.76597134, 51.76737513,
51.76464054, 51.76481595, 51.76542577, 51.76557477, 51.76682149,
51.7644335, 51.76714421, 51.76681267, 51.76597134, 51.76571265,
51.76447255, 51.76339382, 51.76268887, 51.76062289, 51.76030512,
51.76678776, 51.75996884, 51.75968219, 51.75986347, 51.75998767,
51.76749876, 51.76822905, 51.76474771, 51.76863319, 51.76622254,
51.7655237, 51.76482531, 51.76430735, 51.76450781, 51.76421526,
51.76428383, 51.76308822, 51.76434118, 51.76525265, 51.76642077,
51.7672966, 51.76661139, 51.76598088)), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -60L), groups = structure(list(
Time = c(-25, -23, -22, -20, -20, -14, -14, -11, -11, -11,
-10, -10, -10), Code = c(1L, 1L, 1L, 1L, 3L, 1L, 2L, 1L,
2L, 3L, 1L, 2L, 3L), .rows = structure(list(1L, 19:20, 2:4,
22L, 21L, c(7L, 8L, 11L, 12L, 13L, 14L, 15L, 16L, 17L
), c(5L, 6L, 9L, 10L, 18L), 23:30, 32L, 31L, c(33L, 35L,
41L, 42L, 43L, 45L, 47L, 50L, 52L, 53L, 58L, 59L), c(34L,
36L, 37L, 38L, 39L, 40L, 44L, 46L, 49L, 55L, 56L, 60L
), c(48L, 51L, 54L, 57L)), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -13L), .drop = TRUE))
If I was to do this by hand I would use dplyr::filter(Time == "x") then make each plot using leaflet like so
install.packages("leaflet")
library(leaflet)
statecol<- colorFactor(palette = "viridis", df$Code) #create the colour palette
plots<- leaflet() %>% setView(lng = -1.324640, lat = 51.770462, zoom = 13.25)
plots %>% addTiles() %>%
addCircleMarkers(data = df, label = ~as.character(df$Site), radius = 5, color = ~statecol(Code), stroke = FALSE, fillOpacity = 5) %>%
addLegend('bottomright', pal = statecol, values = df$Code,
title = 'Codes',
opacity = 2)
If there is a better solution than a loop I'd be happy to try that too. Hope this is clear and thanks in advance
CodePudding user response:
One approach would be to put the plotting code in a function which takes a the sole argument a dataframe. To make a map for each unique value of Time you could then split your data by Time and loop over the splitted dataset using the plotting function, where instead of a for loop I use lapply. As a result you get a list with plots for each value of Time:
library(leaflet)
library(dplyr)
df_split <- df %>%
ungroup() %>%
split(.$Time)
statecol<- colorFactor(palette = "viridis", df$Code) #create the colour palette
plot_fun <- function(x) {
leaflet() %>%
setView(lng = -1.324640, lat = 51.770462, zoom = 13.25) |>
addTiles() %>%
addCircleMarkers(data = x, label = ~as.character(x$Site), radius = 5, color = ~statecol(Code), stroke = FALSE, fillOpacity = 5) %>%
addLegend('bottomright', pal = statecol, values = x$Code,
title = 'Codes',
opacity = 2)
}
plots <- lapply(df_split, plot_fun)
length(plots)
#> [1] 7
plots[[1]]
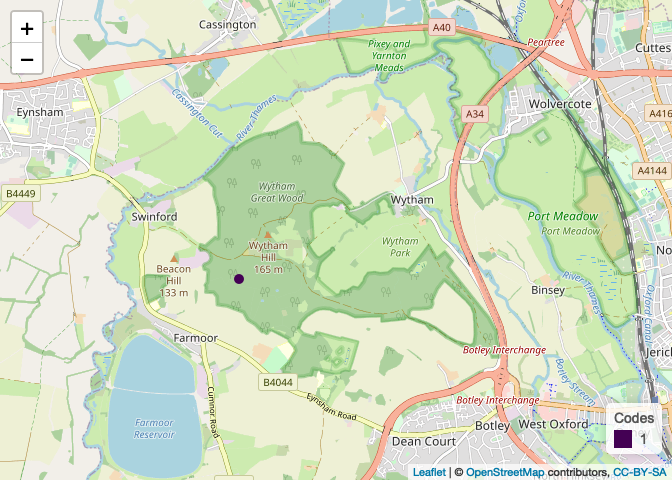
EDIT In case you want to keep or use the data from previous plots we could basically use the same code with one small change, i.e. loop over an index and combine (rbind) the datasets up to the index value inside the plotting function:
library(leaflet)
library(dplyr)
df_split <- df %>%
ungroup() %>%
split(.$Time)
statecol<- colorFactor(palette = "viridis", df$Code) #create the colour palette
plot_fun <- function(ix) {
x <- do.call(rbind, df_split[seq(ix)])
leaflet() %>%
setView(lng = -1.324640, lat = 51.770462, zoom = 13.25) |>
addTiles() %>%
addCircleMarkers(data = x, label = ~as.character(x$Site), radius = 5, color = ~statecol(Code), stroke = FALSE, fillOpacity = 5) %>%
addLegend('bottomright', pal = statecol, values = x$Code,
title = 'Codes',
opacity = 2)
}
plots <- lapply(seq_along(df_split), plot_fun)
plots[[3]]
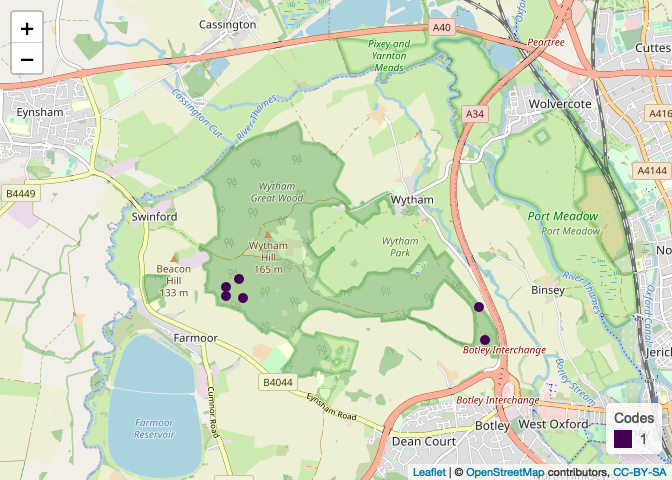
plots[[5]]
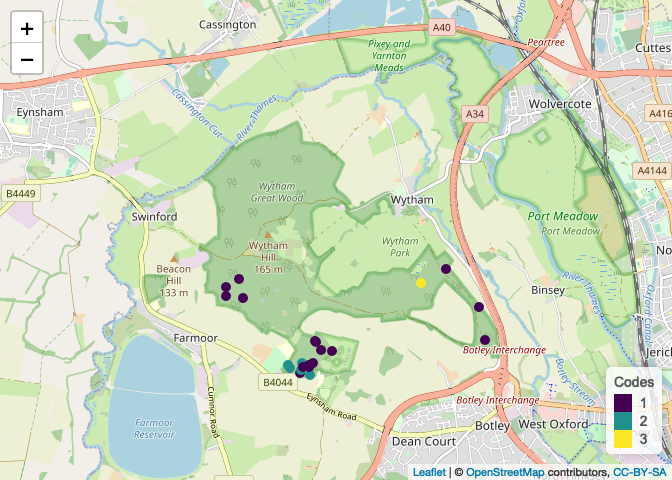
CodePudding user response:
one approach is to store the Time-wise leaflet maps in a column of your tibble:
## create your base map (only needed once):
base_map <- leaflet() %>%
setView(lng = -1.324640, lat = 51.770462, zoom = 13.25) %>%
addTiles()
all_plots <-
df %>% ## df is your tibble given above
group_by(Time) %>%
## compact the data apart from Time as dataframes per Time-group
## into column "nested_data"
nest(nested_data = c(Site, Code, lon, lat)) %>%
rowwise %>%
mutate(leaflet_map = list(
base_map %>%
addCircleMarkers(data = nested_data, ## the column of dataframes
label = ~as.character(Site),
radius = 5,
color = ~statecol(Code),
stroke = FALSE,
fillOpacity = 5
)
)
) %>%
select(Time, leaflet_map)
The resulting tibble can be filtered and iterated (over column leaflet_plot) for automated map display.
## for single map at Time == -25
all_plots %>%
filter(Time == -25) %>%
pull(leaflet_map) %>%
print
Find solutions to display multiple maps in one go here: Multiple leaflets in a grid
