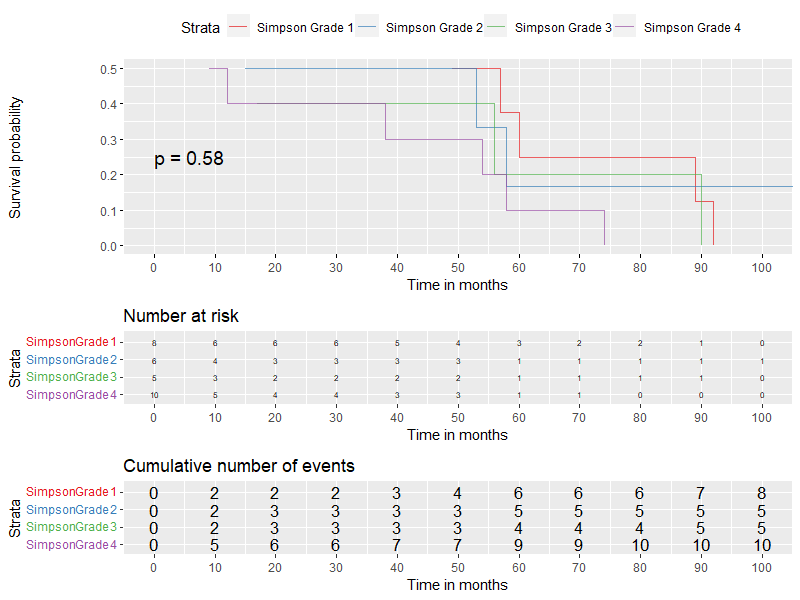
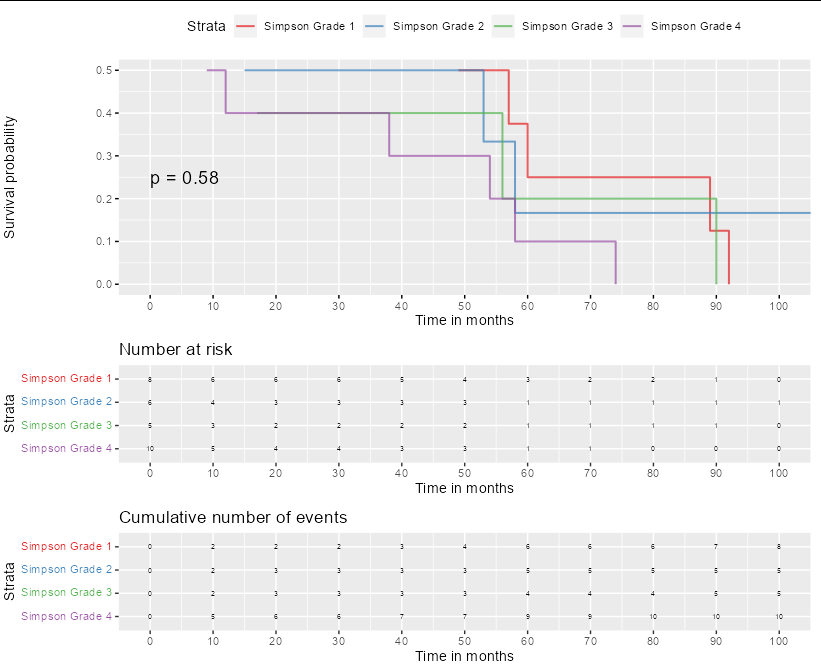
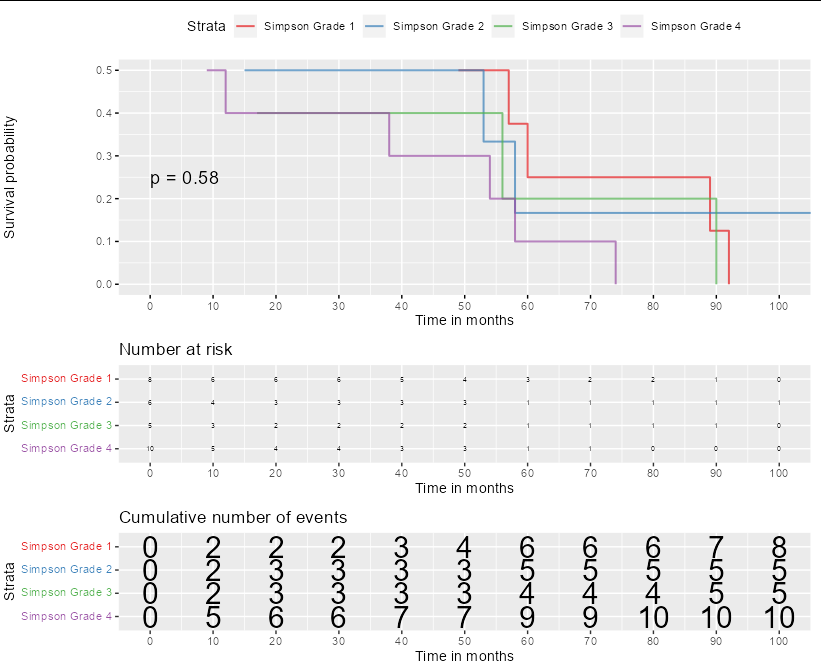
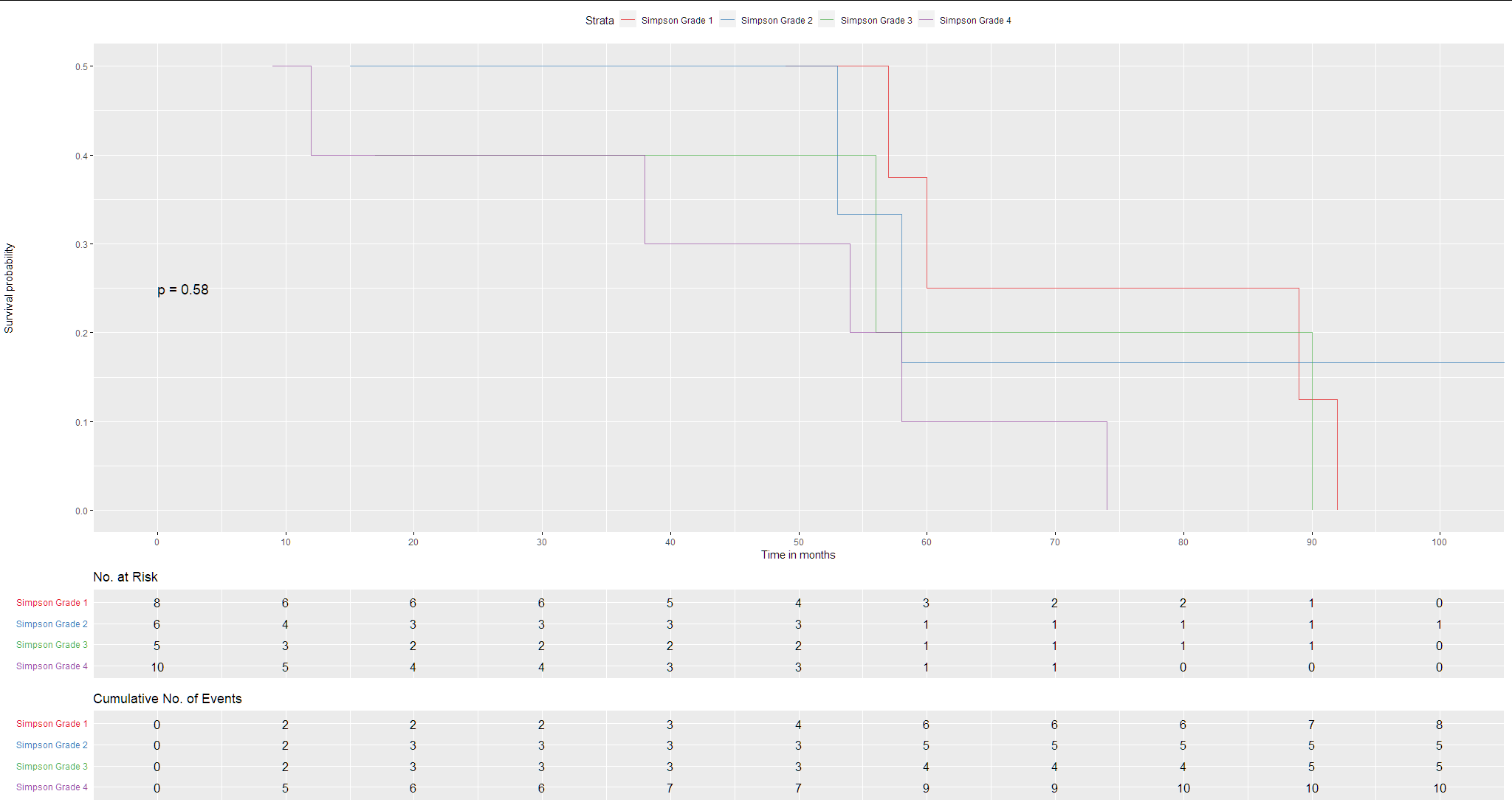
The graph was computed with this
library(survival)
library(survminer)
library(ggplot2)
fit <- survfit(Surv(p$time.recur.months, p$recurrence) ~ p$simpson.grade, conf.type="log", data=p)
ggsurvplot(
fit,
data = p,
risk.table = TRUE,
risk.table.fontsize = 2,
cumevents = TRUE,
cumevents.fontsize =2,
pval = TRUE,
pval.coord = c(0, 0.25),
conf.int = F,
legend.labs=c("Simpson Grade 1" ,"Simpson Grade 2", "Simpson Grade 3",
"Simpson Grade 4"),
size=c(0.7,0.7,0.7,0.7),
xlim = c(0,100),
alpha=c(0.7),
break.time.by = 10,
xlab="Time in months",
#ylab="Survival probability",
ggtheme = theme_gray(),
risk.table.y.text.col = T,
risk.table.y.text = TRUE,
ylim=c(0,0.5),
palette="Set1"
)
My Data
p <- structure(list(recurrence = c(0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L,
0L, 0L, 1L, 0L, 0L, 0L, 1L, 1L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L,
0L, 0L, 1L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L,
0L, 0L, 0L, 0L, 1L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L,
1L, 1L, 0L, 0L, 1L, 1L, 0L, 0L, 0L, 0L, NA, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 1L, 0L, 1L, 0L, 0L, 0L, 1L, 1L, 1L, 1L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 1L, 0L, 0L,
0L, 0L, 0L, 1L, 0L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 1L,
0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L), time.recur.months = c(NA, NA,
NA, NA, NA, NA, 92L, NA, NA, NA, 74L, NA, NA, NA, 2L, 8L, NA,
NA, NA, NA, 58L, NA, NA, NA, NA, NA, 3L, NA, 4L, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, 39L, NA, NA, NA, NA, 15L, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, 12L, 56L, 57L, NA, NA, 49L, 17L,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 5L,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 9L, NA,
89L, NA, NA, NA, 8L, 6L, 8L, 4L, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, 60L, NA, NA, 38L, NA, NA, NA, NA, NA, 90L,
NA, 58L, 54L, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, 53L, NA, NA, 124L, NA, NA,
NA, NA, NA, NA, 7L, NA), simpson.grade = c(3L, 1L, 1L, 2L, 4L,
1L, 1L, 1L, 2L, 1L, 4L, 1L, 1L, 2L, 1L, 2L, 1L, 4L, 2L, 3L, 2L,
1L, 1L, 1L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 1L, 1L, 3L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L,
1L, 1L, 4L, 3L, 1L, 1L, 4L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 3L, 1L,
3L, 4L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 4L, 1L, 1L, 1L, 4L, 1L, 1L,
1L, 2L, 1L, 2L, 4L, 4L, 1L, 4L, 4L, 1L, 2L, 1L, 1L, 4L, 4L, 4L,
4L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 4L, 1L, 2L, 1L, 4L, 1L, 1L, 4L,
4L, 1L, 3L, 1L, 1L, 1L, 3L, 2L, 4L, 4L, 1L, 4L, 4L, 4L, 4L, 1L,
1L, 1L, 1L, 4L, 1L, 4L, 4L, 1L, 4L, 4L, 1L, 4L, 4L, 3L, 1L, 1L,
1L, 4L, 1L, 1L, 1L, 1L, 3L, 1L, 1L, 2L, 2L, 2L, 4L, 1L, 4L, 2L,
1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 3L, 1L)), .Names = c("recurrence",
"time.recur.months", "simpson.grade"), class = "data.frame", row.names
= c(NA,
-176L))
CodePudding user response:
I can't see the documentation for a cumevents.fontsize argument, but a straightforward way to change it is to store the plot and change it directly:
mygg <- ggsurvplot(
fit,
data = p,
risk.table = TRUE,
risk.table.fontsize = 2,
cumevents = TRUE,
cumevents.fontsize =2,
pval = TRUE,
pval.coord = c(0, 0.25),
conf.int = F,
legend.labs=c("Simpson Grade 1" ,"Simpson Grade 2", "Simpson Grade 3",
"Simpson Grade 4"),
size=c(0.7,0.7,0.7,0.7),
xlim = c(0,100),
alpha=c(0.7),
break.time.by = 10,
xlab="Time in months",
#ylab="Survival probability",
ggtheme = theme_gray(),
risk.table.y.text.col = T,
risk.table.y.text = TRUE,
ylim=c(0,0.5),
palette="Set1"
)
So now you can do:
mygg$cumevents$layers[[1]]$aes_params$size <- 2
mygg
or
mygg$cumevents$layers[[1]]$aes_params$size <- 8
mygg
CodePudding user response: