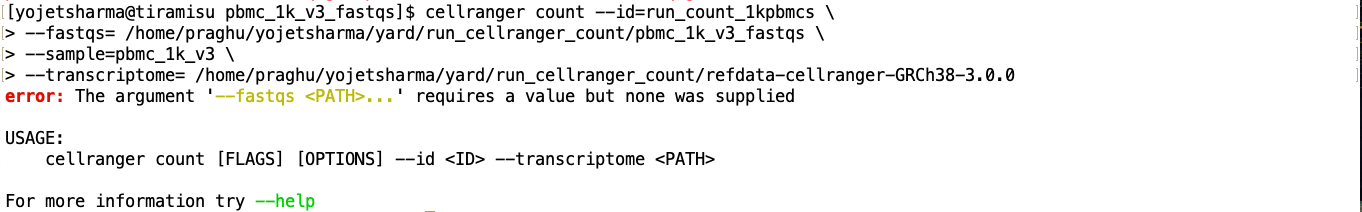
I am following the 10x Cellranger steps and using the same files for cellranger count. I run this from the fastq directory that contains all the PBMCs fastq files and the GRCh38 files too.
CodePudding user response:
You put a space between --fastqs= and /home/.../pbmc_1k_v3_fastqs, and thus it think you didn't provide an argument after --fastqs. There are two possible syntax:
- either you want a space, but then don't put a
=, as in--fastqs /home/.../pbmc_1k_v3_fastqs; - or you put an equal sign, but then don't put a space, as in
--fastqs=/home/.../pbmc_1k_v3_fastqs.
CodePudding user response:
I had the same issue so I suppressed the space but now it is saying that: $ cellranger count --id=run_count_1kpbmcs \
--fastqs=/home/hd/hd_hd/hd_ak282/yard/run_cellranger_count/pbmc_1k_v3_fastqs
--sample=pbmc_1k_v3
--transcriptome=/home/hd/hd_hd/hd_ak282/yard/run_cellranger_count/refdata-cellranger-GRCh38-3.0.0 error: The following required arguments were not provided: --transcriptome
Can someone help me please? :)