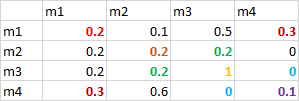
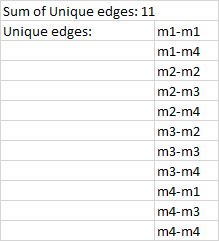
Based on the adjacency matrix, I would like to count the number of unique edges in a network. In the below example I coloured the unique edges between the different nodes. But I don't know how to proceed.
Desired output:
Sample data
structure(list(...1 = c("m1", "m2", "m3", "m4"), m1 = c(0.2,
0.2, 0.2, 0.3), m2 = c(0.1, 0.2, 0.2, 0.6), m3 = c(0.5, 0.2,
1, 0), m4 = c(0.3, 0, 0, 0.1)), row.names = c(NA, -4L), spec = structure(list(
cols = list(...1 = structure(list(), class = c("collector_character",
"collector")), m1 = structure(list(), class = c("collector_double",
"collector")), m2 = structure(list(), class = c("collector_double",
"collector")), m3 = structure(list(), class = c("collector_double",
"collector")), m4 = structure(list(), class = c("collector_double",
"collector"))), default = structure(list(), class = c("collector_guess",
"collector")), delim = ","), class = "col_spec"), class = c("spec_tbl_df",
"tbl_df", "tbl", "data.frame"))
CodePudding user response:
Assuming that this is an undirected graph such that 0 indicates no edge and a positive number indicates an edge, convert the input DF to a logical matrix and from that to an igraph object. Then get its edges and the names of those edges. (Another possible output is by using as_edgelist(g) to get a 2 column matrix such that each row defines an edge.)
library(igraph)
m <- as.matrix(DF[-1])
rownames(m) <- colnames(m)
g <- graph_from_adjacency_matrix(m > 0)
e <- E(g)
attr(e, "vnames")
## [1] "m1|m1" "m1|m2" "m1|m3" "m1|m4" "m2|m1" "m2|m2" "m2|m3" "m3|m1" "m3|m2"
## [10] "m3|m3" "m4|m1" "m4|m2" "m4|m4"
Alternately as a pipeline
library(igraph)
library(tibble)
DF %>%
column_to_rownames("...1") %>%
as.matrix %>%
sign %>%
graph_from_adjacency_matrix %>%
E %>%
attr("vnames")
## [1] "m1|m1" "m1|m2" "m1|m3" "m1|m4" "m2|m1" "m2|m2" "m2|m3" "m3|m1" "m3|m2"
## [10] "m3|m3" "m4|m1" "m4|m2" "m4|m4"
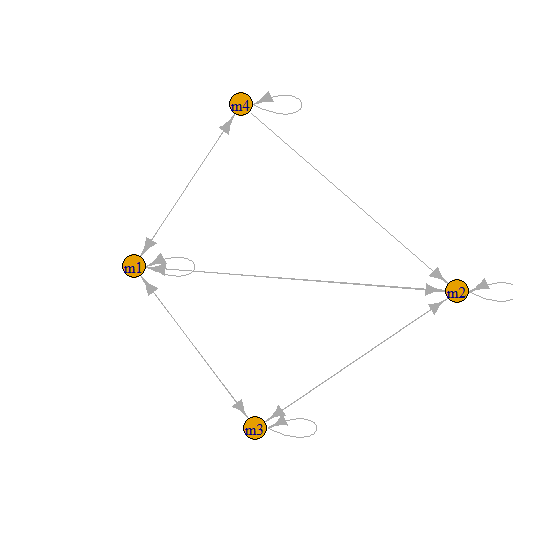
The graph of g looks like this:
set.seed(123)
plot(g)
Note
DF <-
structure(list(...1 = c("m1", "m2", "m3", "m4"), m1 = c(0.2,
0.2, 0.2, 0.3), m2 = c(0.1, 0.2, 0.2, 0.6), m3 = c(0.5, 0.2,
1, 0), m4 = c(0.3, 0, 0, 0.1)), row.names = c(NA, -4L), spec = structure(list(
cols = list(...1 = structure(list(), class = c("collector_character",
"collector")), m1 = structure(list(), class = c("collector_double",
"collector")), m2 = structure(list(), class = c("collector_double",
"collector")), m3 = structure(list(), class = c("collector_double",
"collector")), m4 = structure(list(), class = c("collector_double",
"collector"))), default = structure(list(), class = c("collector_guess",
"collector")), delim = ","), class = "col_spec"), class = c("spec_tbl_df",
"tbl_df", "tbl", "data.frame"))