I made a loop to run all possible combinations of a regression model. As i only need to keep the residuals of the model i made it as such:
df <- data.frame(t(combn(colnames(orig_df), m = 2)))
test = NULL
for (i in 1:(nrow(df)-1)) {
for (j in (i 1):nrow(df)) {
x <- lm(orig_df[[i]] ~ orig_df[[j]], data = orig_df)
y <- x$residuals
test <- cbind(test, y)
}
}
My problem, however, is adding names to the new data frame (here named "test"). I thought a good way to tell the residuals apart would be adding x$call[2].
However if i create the loop so that:
test = NULL
for (i in 1:(nrow(df)-1)) {
for (j in (i 1):nrow(df)) {
x <- lm(orig_df[[i]] ~ orig_df[[j]], data = Telekom)
y <- x$residuals
test <- cbind(test, y)
colnames(test) <- paste(x$call[2])
}
}
After this the loop stops working as it should, and the error displayed is:
length of 'dimnames' [2] not equal to array extent . What is my error here?
Thanks in advance!
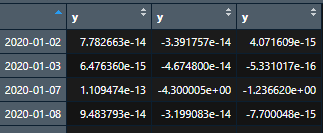
For first loop:
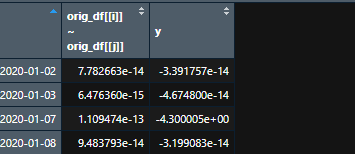
For second loop:
CodePudding user response:
The issue can be reproduced with mtcars data
> model <- lm(mtcars[[1]] ~ mtcars[[2]])
> model$call
lm(formula = mtcars[[1]] ~ mtcars[[2]])
We may assign the call afterwards
> model$call[[2]] <- as.formula(paste0(names(mtcars)[1], "~ ", names(mtcars)[2]))
> model$call
lm(formula = mpg ~ cyl)
In the OP's loop, may be this modification can work
test = NULL
for (i in 1:(nrow(df)-1)) {
for (j in (i 1):nrow(df)) {
x <- lm(orig_df[[i]] ~ orig_df[[j]], data = Telekom)
x$call[[2]] <- as.formula(paste0(names(orig_df)[i], "~ ", names(orig_df)[j])))
y <- x$residuals
test <- cbind(test, y)
colnames(test)[ncol(test)] <- paste(x$call[[2]])
}
}