I'm having trouble trying to iterate through data and combine my columns:
names <- c()
haplotypervalue <- c()
x <- as.numeric(nrow(noNAhaplotypes)) - 1
for (i in 1:x) {
if (haplotypevalue[i] %in% haplotypevalue && i > 2) {
j <- i 1
haplotypevalue[[i]] <- c(haplotypevalue, noNAhaplotypes$value[j])
next
}
names <- c(names, noNAhaplotypes$name[i])
haplotypevalue <- c(haplotypevalue, noNAhaplotypes$value[i])
}
My data looks something like this:
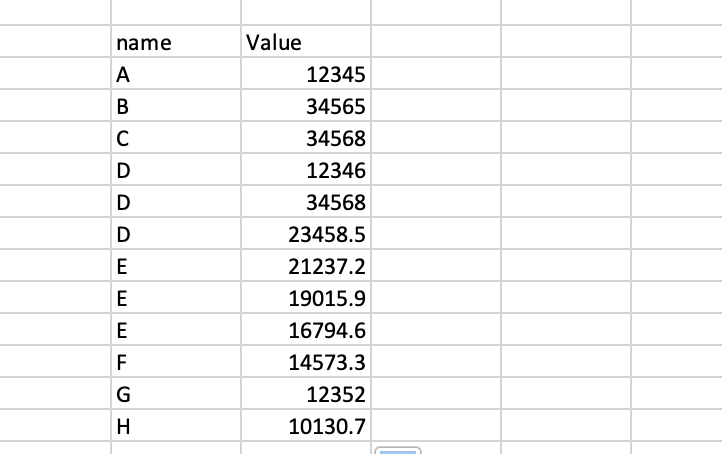
What I am trying to do is effectively loop through the data frame (noNAhaplotypes), and check to see if the 'name' at each row already exists in the name vector I have created. If it does, I want to add corresponding value from the value column to the haplotypevalue vector.
My results should be something like: name: [A, B, C, D...] haplotypevalue: [12345, 34565, 34568, {12346, 34568, 23458.5}...]
This means haplotypevalue[4] should yield all the 3 values associated with 'D'.
I've been trying to do a for loop but I get: " Error in haplotypevalue[[i]] : subscript out of bounds
"
Apologies if the data or variable names are confusing or if some spellings don't match up. I changed the names to protect data integrity.
CodePudding user response:
haplotypervalue <- split(df$value, df$name)
$A
[1] 12345
$B
[1] 34565
$C
[1] 34568
$D
[1] 12346.0 34568.0 23458.5
$E
[1] 21237.2 19015.9 16794.6
$F
[1] 14573.3
$G
[1] 12352
$H
[1] 10130.7
