I am reading a csv file into a pandas dataframe which has duplicate values for the rows and columns that I need to agregate (both rows and cols).
The csv file looks like this:
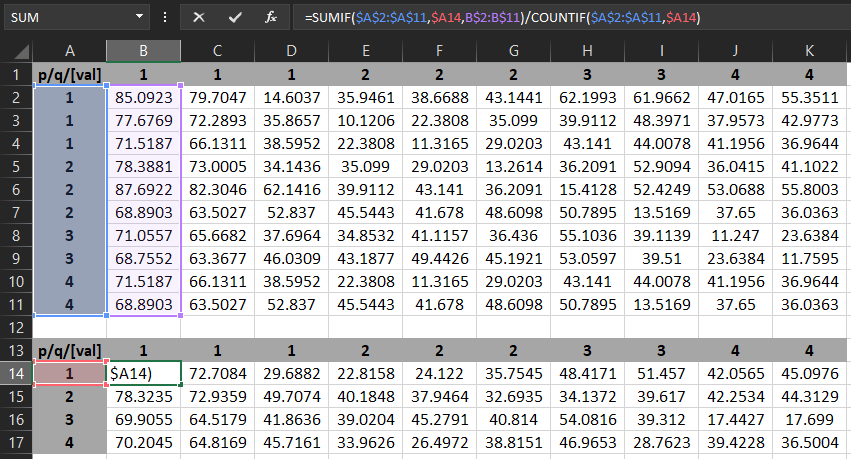
| p/q/[val] | 1 | 1 | 1 | 2 | 2 | 2 | 3 | 3 | 4 | 4 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 85.09227753 | 79.70470428 | 14.60372257 | 35.94606018 | 38.66883087 | 43.14413452 | 62.1992569 | 61.9662056 | 47.01652908 | 55.35105515 |
| 1 | 77.67690277 | 72.28933716 | 35.8657341 | 10.12055206 | 22.38080597 | 35.09898376 | 39.91122818 | 48.39712524 | 37.95729065 | 42.97728348 |
| 1 | 71.51867676 | 66.13111115 | 38.59518433 | 22.38080406 | 11.31649399 | 29.02029228 | 43.14096069 | 44.00777054 | 41.19556427 | 36.96442413 |
| 2 | 78.38805389 | 73.00048828 | 34.14358902 | 35.09897995 | 29.02029228 | 13.26141262 | 36.20913696 | 52.90936279 | 36.04150391 | 41.10220718 |
| 2 | 87.69218445 | 82.30461884 | 62.14162445 | 39.91123581 | 43.14096451 | 36.20913696 | 15.41283798 | 52.42485428 | 53.06882477 | 55.80033112 |
| 2 | 68.89026642 | 63.50270844 | 52.83700562 | 45.54430771 | 41.67800522 | 48.60984421 | 50.78954315 | 13.5169096 | 37.65000153 | 36.0362854 |
| 3 | 71.05574036 | 65.66817474 | 37.6963768 | 34.8531723 | 41.11572266 | 36.43598175 | 55.10356522 | 39.11390305 | 11.24700832 | 23.63844109 |
| 3 | 68.75523376 | 63.36768723 | 46.03090668 | 43.18769836 | 49.4425621 | 45.19208527 | 53.05971527 | 39.51002502 | 23.63843918 | 11.75947094 |
| 4 | 71.51867676 | 66.13111115 | 38.59518433 | 22.38080406 | 11.31649399 | 29.02029228 | 43.14096069 | 44.00777054 | 41.19556427 | 36.96442413 |
| 4 | 68.89026642 | 63.50270844 | 52.83700562 | 45.54430771 | 41.67800522 | 48.60984421 | 50.78954315 | 13.5169096 | 37.65000153 | 36.0362854 |
When I read the csv file into a pandas df, it converts the duplicated column names to decimal numbers and looks like this:
| p/q/[val] | 1 | 1.1 | 1.2 | 2 | 2.1 | 2.2 | 3 | 3.1 | 4 | 4.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 85.09227753 | 79.70470428 | 14.60372257 | 35.94606018 | 38.66883087 | 43.14413452 | 62.1992569 | 61.9662056 | 47.01652908 | 55.35105515 |
| 1 | 77.67690277 | 72.28933716 | 35.8657341 | 10.12055206 | 22.38080597 | 35.09898376 | 39.91122818 | 48.39712524 | 37.95729065 | 42.97728348 |
| 1 | 71.51867676 | 66.13111115 | 38.59518433 | 22.38080406 | 11.31649399 | 29.02029228 | 43.14096069 | 44.00777054 | 41.19556427 | 36.96442413 |
| 2 | 78.38805389 | 73.00048828 | 34.14358902 | 35.09897995 | 29.02029228 | 13.26141262 | 36.20913696 | 52.90936279 | 36.04150391 | 41.10220718 |
| 2 | 87.69218445 | 82.30461884 | 62.14162445 | 39.91123581 | 43.14096451 | 36.20913696 | 15.41283798 | 52.42485428 | 53.06882477 | 55.80033112 |
| 2 | 68.89026642 | 63.50270844 | 52.83700562 | 45.54430771 | 41.67800522 | 48.60984421 | 50.78954315 | 13.5169096 | 37.65000153 | 36.0362854 |
| 3 | 71.05574036 | 65.66817474 | 37.6963768 | 34.8531723 | 41.11572266 | 36.43598175 | 55.10356522 | 39.11390305 | 11.24700832 | 23.63844109 |
| 3 | 68.75523376 | 63.36768723 | 46.03090668 | 43.18769836 | 49.4425621 | 45.19208527 | 53.05971527 | 39.51002502 | 23.63843918 | 11.75947094 |
| 4 | 71.51867676 | 66.13111115 | 38.59518433 | 22.38080406 | 11.31649399 | 29.02029228 | 43.14096069 | 44.00777054 | 41.19556427 | 36.96442413 |
| 4 | 68.89026642 | 63.50270844 | 52.83700562 | 45.54430771 | 41.67800522 | 48.60984421 | 50.78954315 | 13.5169096 | 37.65000153 | 36.0362854 |
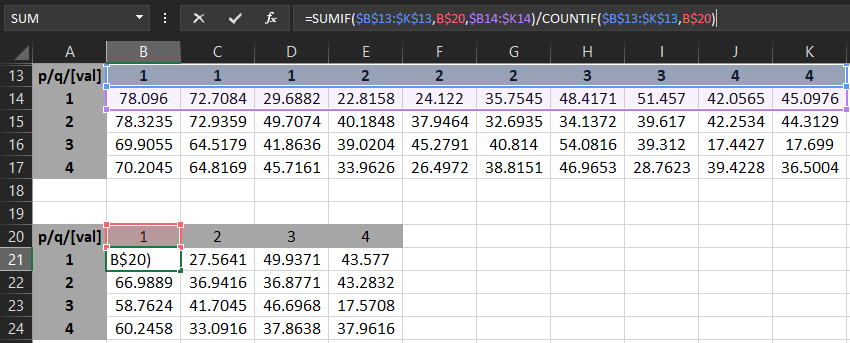
I need to aggregate the rows and the columns so my final dataframe looks like this:
| p/q/[val] | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 60.1641834 | 27.56410641 | 49.93709119 | 43.57702446 |
| 2 | 66.98894882 | 36.94157547 | 36.87710746 | 43.28319232 |
| 3 | 58.76235326 | 41.70453707 | 46.69680214 | 17.57083988 |
| 4 | 60.24582545 | 33.09162458 | 37.863796 | 37.96156883 |
In Excel I can do this in two steps using the following formulae:
Step1 - aggregate the rows:
Step2 - aggregate the columns:
I am just not sure how I can do this in python.
Thanks very much for any help.
CodePudding user response:
If you really have the same indices per column/row:
(df
.set_index('p/q/[val]')
.groupby(level=0).mean()
.groupby(level=0, axis=1).mean()
)
Alternative:
(df
.melt(id_vars='p/q/[val]')
.groupby(['p/q/[val]', 'variable'])['value'].mean()
.unstack()
)
Output:
1 2 3 4
p/q/[val]
1 60.164183 27.564106 49.937091 43.577024
2 66.988949 36.941575 36.877107 43.283192
3 58.762353 41.704537 46.696802 17.570840
4 60.245825 33.091625 37.863796 37.961569
If the columns are on the form 1, 1.1, etc, add a rename step:
(df
.set_index('p/q/[val]')
.rename(columns=lambda x: x.rpartition('.')[0]) # or x[0] if single digits
.groupby(level=0).mean()
.groupby(level=0, axis=1).mean()
)