I have a select request in MySQL that takes between 25-30s, which is extremely long and I was wondering if you could help me fasten it.
CREATE TEMPORARY TABLE results(
id VARCHAR(30),
secondid VARCHAR(5),
allele VARCHAR(30),
translation VARCHAR(10),
level VARCHAR(20),
subgroup VARCHAR(20),
subgroup2 VARCHAR(20)
);
INSERT INTO results(id, secondid, allele, level) SELECT DISTINCT t1.id, t1.secondid, t1.texte, t3.texte
FROM database t1
JOIN database t2 ON t1.id=t2.id
JOIN database t3 ON t1.id=t3.id AND t1.secondid=t3.secondid
WHERE (t1.qualifier,t2.qualifier) = ("allele","organism") AND t3.qualifier = "level_length" AND t3.texte NOT REGEXP "X" AND t3.texte IS NOT NULL
AND t2.texte = ? AND t1.texte REGEXP ?
GROUP BY t1.texte;
UPDATE results SET translation = (SELECT t1.qualifier
FROM database t1
JOIN database t2 ON t1.id=t2.id AND t1.secondid=t2.secondid
JOIN database t3 ON t1.id=t3.id AND t1.secondid=t3.secondid
WHERE t1.qualifier IN ("protein","ncRNA","rRNA") AND t2.texte=results.allele AND t3.texte=results.level LIMIT 1);
UPDATE results SET subgroup = (SELECT t2.subgrp
FROM alleledb.alleleSubgroups t1
JOIN alleledb.subgroups t2 ON t1.subgroup=t2.subgroup
WHERE t1.gene=SUBSTRING_INDEX(results.allele, "*", 1) AND t1.species=? LIMIT 1);
ALTER TABLE results DROP id, DROP secondid;
SELECT * FROM results ORDER BY subgroup ASC, level ASC;
DROP TABLE results;
I need to go through many dbs to get join (same id), database are huge but results to extract are quite low (less than 1% of all the database). The majority of the results are stored in the same db, in different rows (with the same id and secondid). However, id and secondid are not unique to the rows I need to select, only the combinaison of two is.
Thank you.
CodePudding user response:
I would start by having a proper composite index on your database table
First on
(qualifier, id, secondid, texte)
This will help your joins, the where testing and NOT have to go back to the actual raw data tables for the records as the index has the data you are interested in.
Next, I would adjust the query/joins. Since you are specifically looking for the "allele" and "organism" from t1 and t2 respectively, make them as such.
I have no idea what you are doing with your REGEXP "X" or "?" values for texte, but you'll figure that out after.
Here is how I would revise the queries
insert into ...
SELECT DISTINCT
t1.id,
t1.secondid,
t1.texte,
t3.texte
FROM
database t1
JOIN database t2
ON t1.id = t2.id
AND t2.qualifier = 'organism'
JOIN database t3 ON
t1.id = t3.id
AND t1.secondid = t3.secondid
AND t3.qualifier = 'level_length'
WHERE
t1.qualifier = 'allele'
AND t1.texte REGEXP ?
-- I would move these t2 and t3 into the respective JOINs above directly.
AND t3.texte NOT REGEXP "X"
AND t3.texte IS NOT NULL
AND t2.texte = ?
GROUP BY
t1.texte;
As for your UPDATE commands, having a second index on (id, secondid) will help on the join to t2 and t3 since there is no qualifier context to the join.
As for your UPDATE commands, as Rick mentioned, without some context of an ORDER BY clause, you have no guarantee WHICH record is returned back by the LIMIT 1.
CodePudding user response:
First of all, thank you for all your help.
My first table (The insert to and the first update, database named) looks like this :
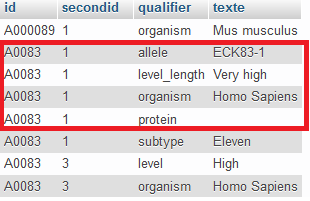
I want all things in red. In others words, I need some parameters which has the same id and secondid as the "level" which is unique among the id. Whereas others parameters may be repeated within the same id (but different second id). I am filtering using the allele name (ECK in EC locus) with thé REGEXP and species. For example, all allèles from EC locus of human.
Then (last update), I take one parameter (allele), substring it and go to a database that gives me one id (one row -> one id). And I use this id on annoter database that gives me one or two rows (one subgroup or two subgroups/rare). So as in my example I only has one group, the absence of ORDER BY was not seen. But yes I want to order (get the subgroup that contains the allele in first). I don't know how to do that.
Finally, I can try to make an index but due to the size of the db, I'm wondering the time and the size of such an index. Would it significally improve time and can I remove it ?
The REGEXP "X" is to remove every matches that are not relevant regarding this parameter (I don't want them). The ? is user input (for the species/2 occurrences this one and the locus).
The operations on the first database takes 30s, last operation on the two databases lasts 1-2s. Others (drop , select) are <20ms (not the problem).
