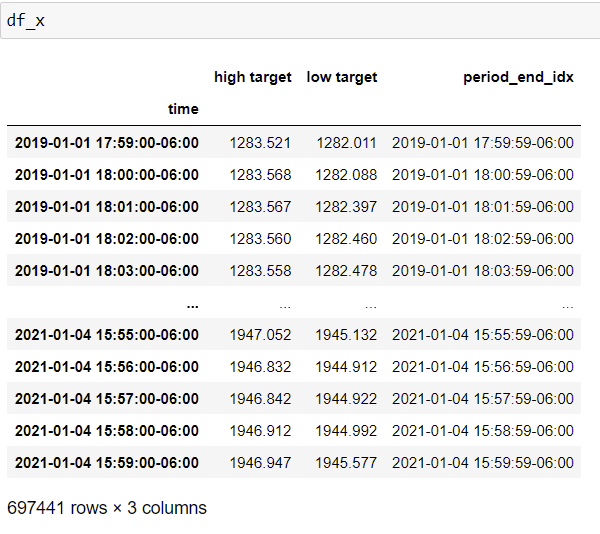
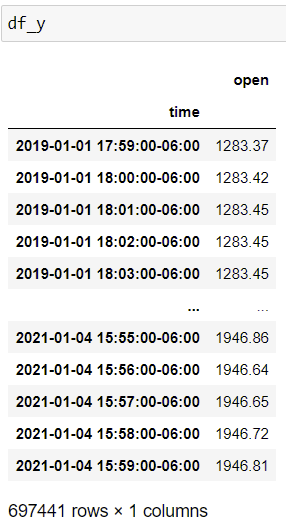
I have two Pandas DataFrames df_x and df_y. df_x has two columns 'high target' and 'low target'. Per every row of df_x, I would like to search through the instances of df_y and see whether the 'high target' was reached before the 'low target'. Currently, I implemented the above using .apply. However, my code is too inefficient as it linearly scales with the number of rows in df_x. Any suggestions to optimize/vectorize my code?
def efficient_high_after_low(row, minute_df):
"""True, if high happened after the low, else False.
Args:
row: A pandas dataframe
with high, low,
minute_df: the whole dataframe
"""
minute_df_after = minute_df.loc[row.period_end_idx pd.Timedelta(minutes=1):]
#print(minute_df_after)
first_highs = (minute_df_after.ge(row['high target']))
first_lows = (minute_df_after.le(row['low target']))
hi_sum, lo_sum = first_highs.sum(), first_lows.sum()
if (len(first_highs) != len(first_lows)):
raise Exception('Unequal length of first_highs and first_lows')
else:
if ((len(first_highs) == 0)):
return None
elif ((hi_sum == 0) & (lo_sum != 0)):
return True
elif ((hi_sum != 0) & (low_sum == 0)):
return False
elif ((hi_sum == 0) & (low_sum == 0)):
return None
elif (first_highs.idxmax() > first_lows.idxmax()):
return True
elif(first_highs.idxmax() < first_lows.idxmax()):
return False
else:
return None
And I do the following to get these boolean values:
df_x.apply(efficient_high_after_low, axis=1, args=(df_y['open'],))
Running the code above on the first 1000 lines takes 4 seconds.
CodePudding user response:
Is this totally off? I dont have your dataframe to test it, but I would think it does what you want.
df_x = pd.DataFrame(np.random.randint(low=-100,high=500,size=500000), columns=["open"], index=[str(i) for i in range(500000)])
df_x["open"] = df_x["open"].cumsum()/100000
df_x["high target"] = df_x["open"] 0.1
df_x["low target"] = df_x["open"]-0.1
# This is just a place holder which keeps .sum() count correct
df_x["low_first"] = 0
for nrow in range(len(df_x.index)):
# How does Open continue nrow steps in to the future
df_x["nrow"] = df_x["open"].shift(-nrow)
if df_x["low_first"].abs().sum()>=df_x["nrow"].notna().sum(): break #No needless searching
# Did we break our high thresh hold?
df_x["low_first"] = df_x["low_first"] (df_x["high target"] <= df_x["nrow"])*(1-df_x["low_first"].abs())
# Did we break our low thresh hold?
df_x["low_first"] = df_x["low_first"] (df_x["low target"] >= df_x["nrow"])*(df_x["low_first"].abs()-1)
# Cleaning up
df_x = df_x.drop(columns=["nrow"])
# And if you want your low/high columns
df_x["first_high"] = df_x["low_first"].replace(-1,0)
df_x["first_low"] = df_x["low_first"].replace(1,0)*(-1)
CodePudding user response:
This is what you could do:
First of all put the open column in your main dataframe, let's call it df (note: this only works if you have the exact same index on df_y, if you don't, consider other solutions like pd.concat or pd.merge_asof)
df = df_x
df["open"] = df_y["open"]
I also took the liberty of renaming your columns.
As long as your timeseries index is ordered, we can reset the index with
df = df.reset_index()
So now we have df something like this (values are made up):
high_trgt low_trgt open
0 8.746911 8.712824 9.243329
1 9.472977 10.190079 9.744083
2 9.445111 10.269676 9.859353
3 9.972061 10.014381 9.132204
4 8.934692 8.914729 11.453276
# You "time" column isn't actually necessary for this solution
We can create a map of where the targets have been hit
map_high = df.open.values > df.high_trgt.values
map_low = df.open.values > df.low_trgt.values
Now the resource intensive bit:
df["high_was_hit_on"] = pd.Series([map_high[i:].argmax() for i in range(len(map_high))])
df["low_was_hit_on"] = pd.Series([map_low[i:].argmax() for i in range(len(map_low))])
Output:
high_trgt low_trgt open high_was_hit_on low_was_hit_on
0 8.746911 8.712824 9.243329 0 0
1 9.472977 10.190079 9.744083 0 3
2 9.445111 10.269676 9.859353 0 2
3 9.972061 10.014381 9.132204 1 1
4 8.934692 8.914729 11.453276 0 0
What I did here is iterating over the range of the columns, and just checking where the highest value (hence, index) starting from that row is in the map that we created before.
Now we can easily check which happened first by doing:
# Here you can customize what you need the results to be
# when two hits happen at the same time
df["high_after_low"] = df.high_was_hit_on < df.low_was_hit_on
In terms of speed, this is the test over a df with 1M rows:
%timeit find_first_hit(df)
3.46 s ± 253 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Truth be told, this isn't exactly vectorized, but I can't think of anything that you could do to really achieve that here. Hope that my solution is helpful anyway.