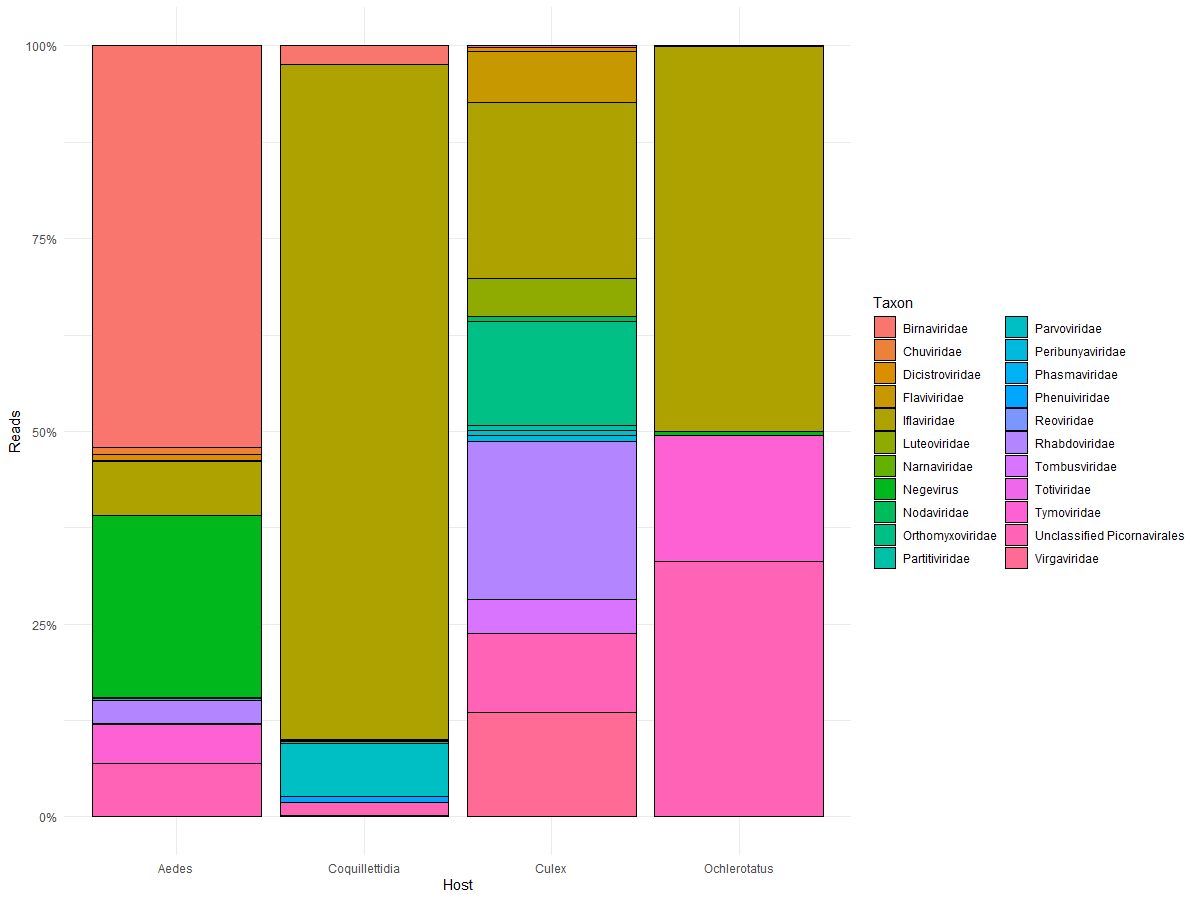
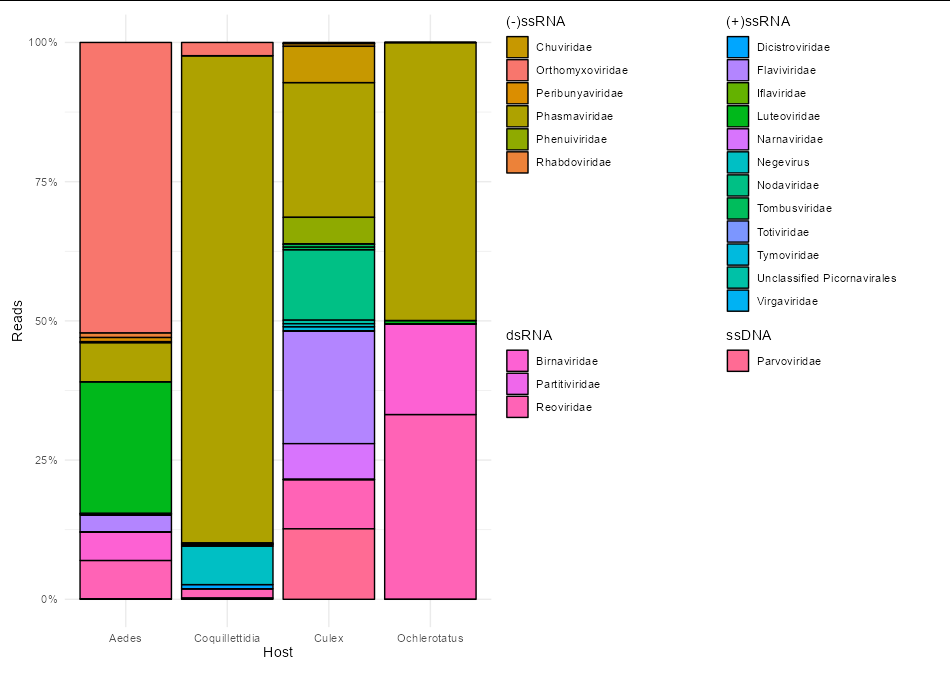
So I have this bar chart I made using:
ggplot(ViralReads, aes(x=Host, y=Reads, fill=Taxon))
geom_col(colour = "black", position = "fill")
scale_y_continuous(labels = label_percent())
theme_minimal()
For each "taxon" there is a category 'Genome' associated with it. I would like to group the items in the legend by 'Genome'. Anyone know if this is possible to do? I tried to play around with the legend a
Here is reproducible data as requested!
structure(list(Host = c("Culex", "Culex", "Culex", "Culex", "Culex",
"Culex", "Culex", "Culex", "Culex", "Culex", "Culex", "Culex",
"Culex", "Culex", "Culex", "Culex", "Culex", "Culex", "Culex",
"Culex", "Culex", "Culex", "Culex", "Aedes", "Aedes", "Aedes",
"Aedes", "Aedes", "Aedes", "Aedes", "Aedes", "Aedes", "Aedes",
"Aedes", "Aedes", "Aedes", "Aedes", "Aedes", "Aedes", "Aedes",
"Aedes", "Aedes", "Aedes", "Aedes", "Aedes", "Aedes", "Ochlerotatus",
"Ochlerotatus", "Ochlerotatus", "Ochlerotatus", "Ochlerotatus",
"Ochlerotatus", "Ochlerotatus", "Ochlerotatus", "Ochlerotatus",
"Ochlerotatus", "Ochlerotatus", "Ochlerotatus", "Ochlerotatus",
"Ochlerotatus", "Ochlerotatus", "Ochlerotatus", "Ochlerotatus",
"Ochlerotatus", "Ochlerotatus", "Ochlerotatus", "Ochlerotatus",
"Ochlerotatus", "Ochlerotatus", "Coquillettidia", "Coquillettidia",
"Coquillettidia", "Coquillettidia", "Coquillettidia", "Coquillettidia",
"Coquillettidia", "Coquillettidia", "Coquillettidia", "Coquillettidia",
"Coquillettidia", "Coquillettidia", "Coquillettidia", "Coquillettidia",
"Coquillettidia", "Coquillettidia", "Coquillettidia", "Coquillettidia",
"Coquillettidia", "Coquillettidia", "Coquillettidia", "Coquillettidia",
"Coquillettidia"), Genome = c("(-)ssRNA", "(-)ssRNA", "(-)ssRNA",
"(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "dsRNA", "dsRNA",
"dsRNA", "ssDNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA",
"(-)ssRNA", "(-)ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "dsRNA", "dsRNA", "dsRNA",
"ssDNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA",
"(-)ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "dsRNA", "dsRNA", "dsRNA", "ssDNA", "(-)ssRNA",
"(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "(-)ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA", "( )ssRNA",
"dsRNA", "dsRNA", "dsRNA", "ssDNA"), Reads = c(102317, 163616,
6188, 130, 0, 0, 195632, 38928, 51823, 4033, 71500, 0, 0, 102317,
3953, 363, 52677, 62, 4033, 5183, 1542, 0, 4313, 1071, 36260,
0, 10089, 0, 0, 84154, 0, 814, 0, 82223, 281910, 60584, 1071,
8802, 0, 2203, 0, 0, 0, 622680, 0, 2229, 0, 0, 0, 550, 0, 0,
1445303, 0, 0, 0, 960293, 16515, 470781, 0, 0, 701, 0, 0, 0,
0, 0, 0, 0, 352, 0, 0, 0, 0, 1186, 138498, 200, 0, 197, 2640,
0, 0, 352, 0, 0, 0, 0, 197, 0, 3752, 0, 10899), Taxon = c("Orthomyxoviridae",
"Rhabdoviridae", "Peribunyaviridae", "Chuviridae", "Phasmaviridae",
"Phenuiviridae", "Iflaviridae", "Luteoviridae", "Tombusviridae",
"Nodaviridae", "Unclassified Picornavirales", "Negevirus", "Tymoviridae",
"Virgaviridae", "Dicistroviridae", "Totiviridae", "Flaviviridae",
"Narnaviridae", "Nodaviridae", "Partitiviridae", "Birnaviridae",
"Reoviridae", "Parvoviridae", "Orthomyxoviridae", "Rhabdoviridae",
"Peribunyaviridae", "Chuviridae", "Phasmaviridae", "Phenuiviridae",
"Iflaviridae", "Luteoviridae", "Tombusviridae", "Nodaviridae",
"Unclassified Picornavirales", "Negevirus", "Tymoviridae", "Virgaviridae",
"Dicistroviridae", "Totiviridae", "Flaviviridae", "Narnaviridae",
"Nodaviridae", "Partitiviridae", "Birnaviridae", "Reoviridae",
"Parvoviridae", "Orthomyxoviridae", "Rhabdoviridae", "Peribunyaviridae",
"Chuviridae", "Phasmaviridae", "Phenuiviridae", "Iflaviridae",
"Luteoviridae", "Tombusviridae", "Nodaviridae", "Unclassified Picornavirales",
"Negevirus", "Tymoviridae", "Virgaviridae", "Dicistroviridae",
"Totiviridae", "Flaviviridae", "Narnaviridae", "Nodaviridae",
"Partitiviridae", "Birnaviridae", "Reoviridae", "Parvoviridae",
"Orthomyxoviridae", "Rhabdoviridae", "Peribunyaviridae", "Chuviridae",
"Phasmaviridae", "Phenuiviridae", "Iflaviridae", "Luteoviridae",
"Tombusviridae", "Nodaviridae", "Unclassified Picornavirales",
"Negevirus", "Tymoviridae", "Virgaviridae", "Dicistroviridae",
"Totiviridae", "Flaviviridae", "Narnaviridae", "Nodaviridae",
"Partitiviridae", "Birnaviridae", "Reoviridae", "Parvoviridae"
)), row.names = c(NA, -92L), class = c("tbl_df", "tbl", "data.frame"
))
CodePudding user response:
The short answer here is "no", you can't have grouped legends within ggplot natively.
However, the long answer is "yes, but it isn't easy". It requires creating a bunch of plots (one per genome) and harvesting their legends, then stitching them back onto the main plot:
library(tidyverse)
fill_df <- ViralReads %>%
select(-1, -3) %>%
unique() %>%
mutate(color = scales::hue_pal()(22))
legends <- lapply(split(ViralReads, ViralReads$Genome), function(x) {
genome <- x$Genome[1]
patchwork::wrap_elements(full = cowplot::get_legend(
ggplot(x, aes(Host, Reads, fill = Taxon))
geom_col(color = "black")
scale_fill_manual(
name = genome,
values = setNames(fill_df$color[fill_df$Genome == genome],
fill_df$Taxon[fill_df$Genome == genome]))
theme(legend.justification = c(0, 1))))
})
p1 <- ggplot(ViralReads, aes(x=Host, y=Reads, fill=Taxon))
geom_col(colour = "black", position = "fill")
scale_y_continuous(labels = scales::label_percent())
theme_minimal()
theme(legend.position = "none")
p1 ((legends[[1]] legends[[2]]) / (legends[[3]] legends[[4]]))
Unfortunately, 22 different colors is just too many to use as a discrete color scale. I would probably use a different way of visualising these data. Anyway, it's good to know that grouped legends are at least possible in ggplot, so thanks for the OP.