I want to find the most important features in my model using shap.
I have this code:
from sklearn.model_selection import train_test_split
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import KFold
import shap
import pandas as pd
import numpy as np
#loading and preparing the data
iris = load_breast_cancer()
X = iris.data
y = iris.target
columns = iris.feature_names
#if you don't shuffle you wont need to keep track of test_index, but I think
#it is always good practice to shuffle your data
kf = KFold(n_splits=2,shuffle=True)
list_shap_values = list()
list_test_sets = list()
for train_index, test_index in kf.split(X):
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
X_train = pd.DataFrame(X_train,columns=columns)
X_test = pd.DataFrame(X_test,columns=columns)
#training model
clf = RandomForestClassifier(random_state=0)
clf.fit(X_train, y_train)
#explaining model
explainer = shap.TreeExplainer(clf)
shap_values = explainer.shap_values(X_test)
#for each iteration we save the test_set index and the shap_values
list_shap_values.append(shap_values)
list_test_sets.append(test_index)
#combining results from all iterations
test_set = list_test_sets[0]
shap_values = np.array(list_shap_values[0])
for i in range(1,len(list_test_sets)):
test_set = np.concatenate((test_set,list_test_sets[i]),axis=0)
shap_values = np.concatenate((shap_values,np.array(list_shap_values[i])),axis=1)
#bringing back variable names
X_test = pd.DataFrame(X[test_set],columns=columns)
#creating explanation plot for the whole experiment, the first dimension from shap_values indicate the class we are predicting (0=0, 1=1)
#shap.summary_plot(shap_values[1], X_test)
shap_sum = np.abs(shap_values).mean(axis=0)
#columns = full_X_train.columns
X_test = pd.DataFrame(X[test_set],columns=columns)
importance_df = pd.DataFrame([X_test.columns.tolist(),shap_sum.tolist()]).T
importance_df.columns = ['column_name','shap_importance']
importance_df = importance_df.sort_values('shap_importance',ascending=False)
print(importance_df)
The output is:
390 None [0.07973283098297632, 0.012745693741197047, 0....
477 None [0.07639585953247056, 0.012705549054148915, 0....
542 None [0.07263038600009886, 0.004509187889530952, 0....
359 None [0.07006782821092902, 0.008022265024270826, 0....
292 None [0.06501143916982145, 0.014648801487419996, 0....
.. ... ...
129 None [0.001207252383050206, 0.005154096692481416, 0...
68 None [0.000537261423323933, 0.000554437257101772, 0...
229 None [0.00046312350178067416, 0.0171676941721087, 0...
94 None [0.00016002701188627102, 0.015384623641506117,...
97 None [0.0001434577248065334, 0.01162161896706629, 0...
This isn't correct, the column names are all None, and it's not clear to me what the shap values are (I was expecting one number for each column ranked from most important features at the top of what's being printed - not a list).
I was hoping for something more like:
Column Shap value
Age 0.3
Gender 0.2
Could someone show me where I went wrong, and how to list the important features for my model using this method?
CodePudding user response:
If you check:
shap_values.shape
(2, 569, 30)
and
X_test.shape
(569, 30)
you'll find out a surprising coincidence on the last 2 axes, which happened not by chance:
- The first axis in shap values is for classes
- Second stands for the number of datapoints in the dataset you iterated over
- Third one is for the actual shap values.
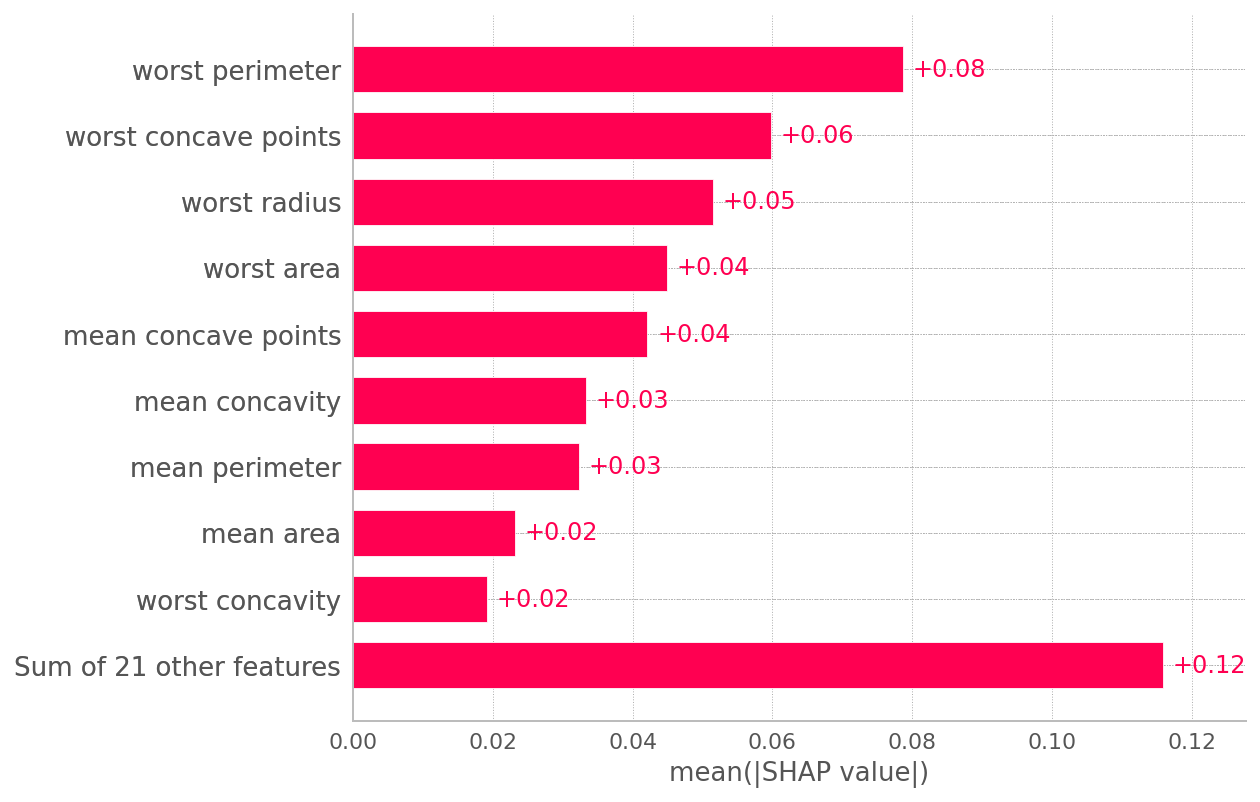
Then, asking the question "what, on average, are the most influential features judged by Shapley contributions" you'll get:
sv = np.abs(shap_values[1,:,:]).mean(0)
cols = X_test.columns
importance_df = pd.DataFrame({
"column_name": cols,
"shap_values": sv
})
#expected result
importance_df.sort_values("shap_values", ascending=False)
column_name shap_values
22 worst perimeter 0.078686
27 worst concave points 0.059845
20 worst radius 0.051551
23 worst area 0.044879
7 mean concave points 0.042114
6 mean concavity 0.033258
2 mean perimeter 0.032346
...
Same as:
shap.plots.bar(shap.Explanation(shap_values[1], feature_names=X_test.columns))