I'm creating a stacked bar plot of relative abundance data, but I only want to display selected few interesting taxa in the legend. I have tried using scale_fill_manual(values = sample(col_vector), breaks = legend_list). The result only shows selected legend in my legend_list as I wish, but all other factors shows no color. How do I show all the colors as stacked bar plot, but only show legend for factors in legend_list?
My code:
ggplot(df, aes_string(x = x, y = y, fill = fill))
geom_bar(stat="identity", position="stack")
scale_fill_manual(values = sample(col_vector),
breaks = legend_list)
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
CodePudding user response:
The reason for your issue is most likely that you are using an unnamed vector of colors. Under the hood, when you pass a vector to the breaks argument this vector is used as names for the color vector passed to the values argument. However, when the number of breaks is smaller than the number of colors only some colors get named while the names for all other colors are set to NA. As a consequence only the categories specified via the breaks argument are assigned a fill color while all other categories are assigned the na.value which by default is "grey50".
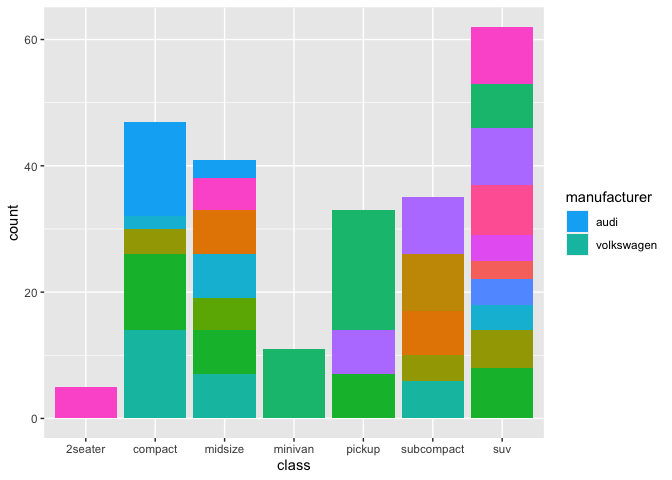
As your provided no minimal reproducible example I use a basic example based on the ggplot2::mpg dataset to first try to reproduce you issue before offering a fix:
library(ggplot2)
library(dplyr)
base <- ggplot(mpg, aes(class, fill = manufacturer))
geom_bar()
legend_list <- c("audi", "volkswagen")
col_vector <- scales::hue_pal()(n_distinct(mpg$manufacturer))
base scale_fill_manual(values = sample(col_vector), breaks = legend_list)
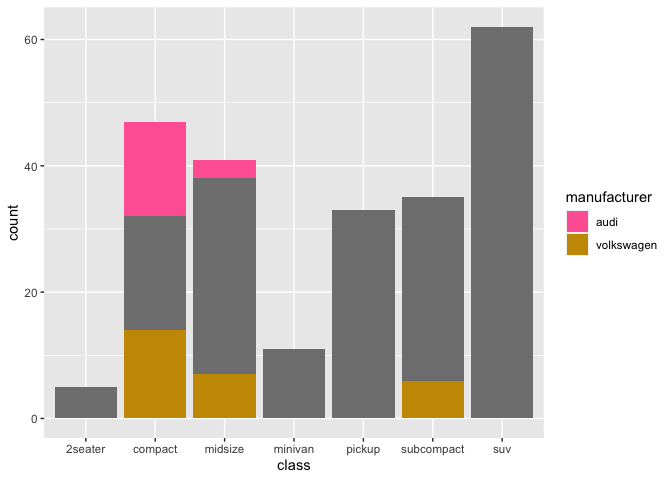
One option to fix the issue is to use a named vector of colors. Doing so all categories are assigned a color but only the categories specified via the breaks argument will show up in the legend:
names(col_vector) <- sample(unique(mpg$manufacturer))
base scale_fill_manual(values = col_vector, breaks = legend_list)