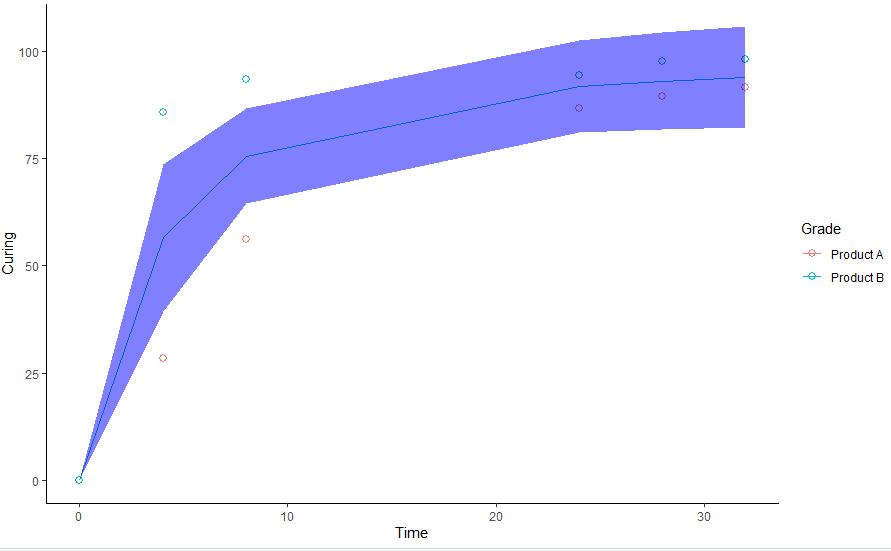
I am willing to interpolate two curves, according to the function Curing ~ a * atan(b * Time), fitting the data reported in the code below. I am getting two problems with this:
library(tidyverse)
library(investr)
library(ggplot2)
#DATAFRAME
RawData <- data.frame("Time" = c(0, 4, 8, 24, 28, 32, 0, 4, 8, 24, 28, 32), "Curing" = c(0, 28.57, 56.19, 86.67, 89.52, 91.42, 0, 85.71, 93.33, 94.28, 97.62, 98.09), "Grade" = c("Product A", "Product A", "Product A", "Product A", "Product A", "Product A", "Product B", "Product B", "Product B", "Product B", "Product B", "Product B"))
attach(RawData)
model <- nls(Curing ~ a * atan(b * Time), data= RawData, control=nls.control(printEval=TRUE, minFactor=2^-24, warnOnly=TRUE))
new.data <- data.frame(time=seq(1, 32, by = 0.1))
interval <- as_tibble(predFit(model, newdata = new.data, interval = "confidence", level= 0.9)) %>% mutate(Time = RawData$Time)
The first is an error as soon as I input the last line:
Error in assign(xname, newdata[, xname]) : first argument not valid
I have tried to change the values of new.data without success. If I remove the optional argument newdata = I can fit, but it looks like the fitting is made interpolating the whole set of data without differentiating the two series.
Below the command lines for getting the graph:
Graph <- ggplot(data=RawData, aes(x=`Time`, y=`Curing`, col=Grade)) geom_point(aes(color = Grade), shape = 1, size = 2.5)
Graph geom_line(data=interval, aes(x = Time, y = fit))
geom_ribbon(data=interval, aes(x=Time, ymin=lwr, ymax=upr), alpha=0.5, inherit.aes=F, fill="blue")
theme_classic()
Is it possible to have both: a smooth and series-separated fitting?
CodePudding user response:
Your error is caused by a typo (time instead of Time in new.data). However, this will not fix the problem of getting one ribbon for each series.
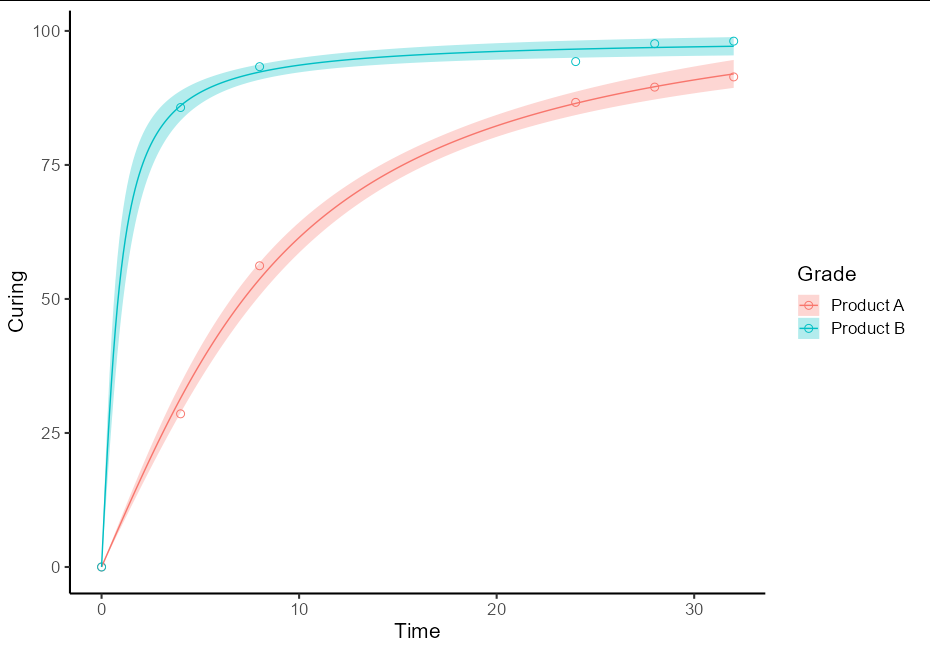
To do this as a one-off, you will need two separate models for the two different sets of data. It is best to use the split-apply-bind idiom to create a single prediction data frame. It also helps plotting if this has a Grade column and the fit column is renamed to Curing
library(tidyverse)
library(investr)
library(ggplot2)
pred_df <- do.call(rbind, lapply(split(RawData, RawData$Grade), function(d) {
new.data <- data.frame(Time = seq(0, 32, by = 0.1))
nls(Curing ~ a * atan(b * Time), data = d, start = list(a = 5, b = 1)) %>%
predFit(newdata = new.data, interval = "confidence", level = 0.9) %>%
as_tibble() %>%
mutate(Time = new.data$Time,
Grade = d$Grade[1],
Curing = fit)
}))
This then allows the plot to be quite straightforward:
ggplot(data = RawData, aes(x = Time, y = Curing, color = Grade))
geom_point(shape = 1, size = 2.5)
geom_ribbon(data = pred_df, aes(ymin = lwr, ymax = upr, fill = Grade),
alpha = 0.3, color = NA)
geom_line(data = pred_df)
theme_classic(base_size = 16)
General approach
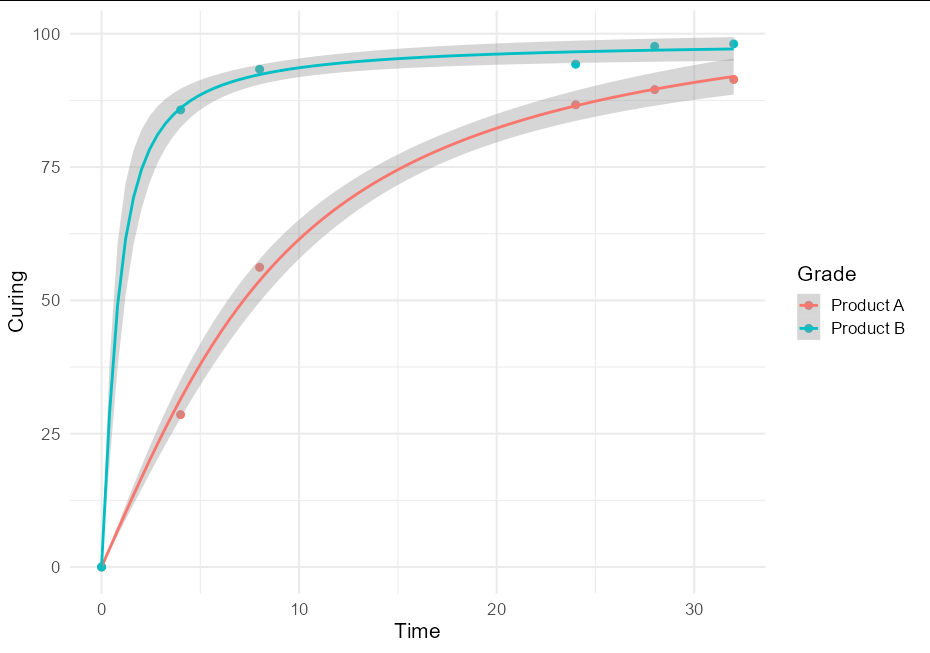
I think this is quite a useful technique, and might be of broader interest, so a more general solution if one wishes to plot confidence bands with an nls model using geom_smooth would be to create little wrappers around nls and predFit:
nls_se <- function(formula, data, start, ...) {
mod <- nls(formula, data, start)
class(mod) <- "nls_se"
mod
}
predict.nls_se <- function(model, newdata, level = 0.9, ...) {
class(model) <- "nls"
p <- investr::predFit(model, newdata = newdata,
interval = "confidence", level = level)
list(fit = p, se.fit = p[,3] - p[,1])
}
This allows very simple plotting with ggplot:
ggplot(data = RawData, aes(x = Time, y = Curing, color = Grade))
geom_point(size = 2.5)
geom_smooth(method = nls_se, formula = y ~ a * atan(b * x),
method.args = list(start = list(a = 5, b = 1)))
theme_minimal(base_size = 16)
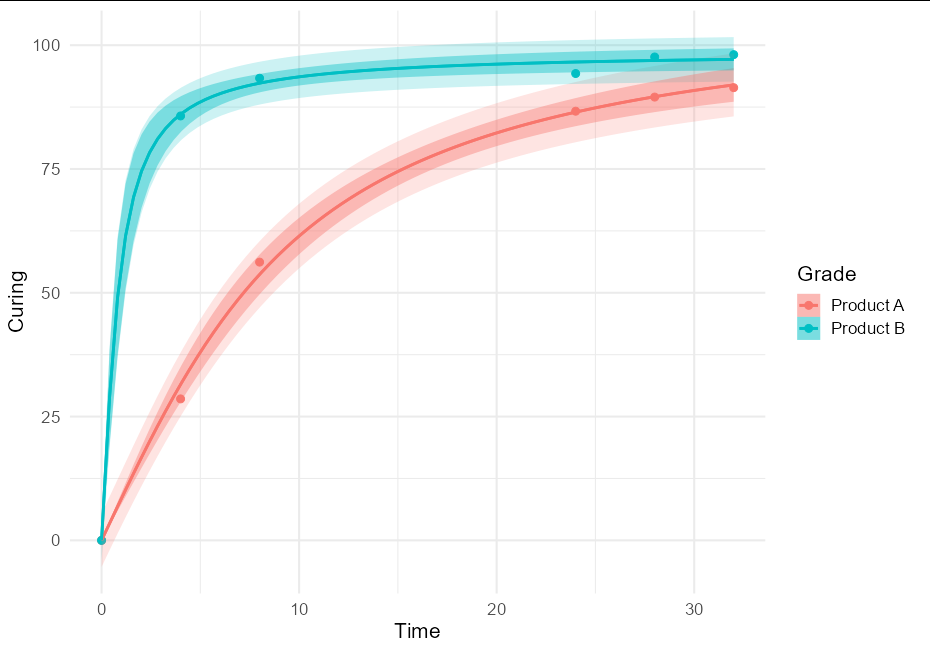
To put both prediction and confidence bands, we can do:
nls_se <- function(formula, data, start, type = "confidence", ...) {
mod <- nls(formula, data, start)
class(mod) <- "nls_se"
attr(mod, "type") <- type
mod
}
predict.nls_se <- function(model, newdata, level = 0.9, interval, ...) {
class(model) <- "nls"
p <- investr::predFit(model, newdata = newdata,
interval = attr(model, "type"), level = level)
list(fit = p, se.fit = p[,3] - p[,1])
}
ggplot(data = RawData, aes(x = Time, y = Curing, color = Grade))
geom_point(size = 2.5)
geom_smooth(method = nls_se, formula = y ~ a * atan(b * x),
method.args = list(start = list(a = 5, b = 1),
type = "prediction"), alpha = 0.2,
aes(fill = after_scale(color)))
geom_smooth(method = nls_se, formula = y ~ a * atan(b * x),
method.args = list(start = list(a = 5, b = 1)),
aes(fill = after_scale(color)))
theme_minimal(base_size = 16)