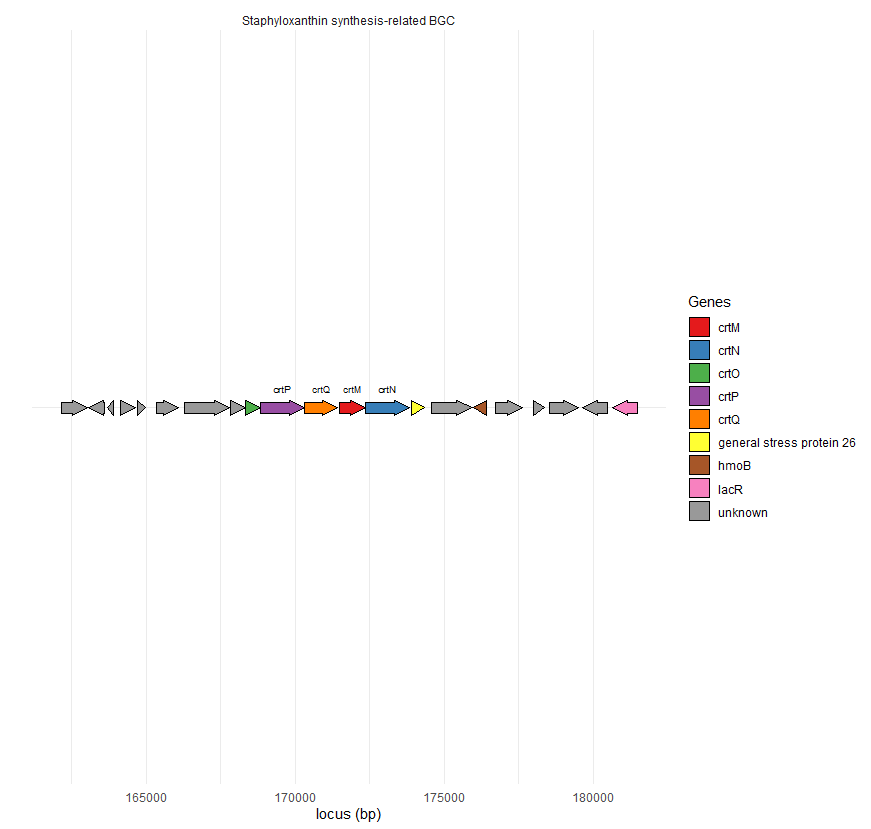
So I'm representing a gene cluster ith the help of the package gggenes. Struggled to find info online but I'm getting there. I started by creating the dataframe with the info:
Molecule <- rep("Staphyloxanthin synthesis-related BGC", 21)
start1 <- c(162155,163052,163705,164135,164702,165358,166305,167847,168352,168826,170323,171480,172352,
173908,174568,175973,176730,178006,178528,179646,180648)
end1 <- c(163024,163609,163911,164641,164977,166086,167804,168347,168825,170316,171426,172355,173857,
174333,175941,176410,177626,178377,179523,180476,181487)
Genes <- c("unknown","unknown","unknown","unknown","unknown","unknown","unknown","unknown","crtO","crtP","crtQ","crtM","crtN","general stress protein 26","unknown","hmoB","unknown","unknown","unknown","unknown","lacR")
strand1 <- c("forward","reverse","reverse","forward","forward","forward","forward","forward","forward","forward","forward","forward","forward","reverse","forward","forward","forward","reverse","reverse","forward","reverse")
orientation1 <- c(1,-1,-1,1,1,1,1,1,1,1,1,1,1,1,1,-1,1,1,1,-1,-1)
BGC1 <- data.frame(Molecule, start1, end1, Genes, strand1, orientation1)
BGC1
Then created a lil vector because I only want to show some relevant genes, not all:
C1 <- c("","","","","","","","","crtO","crtP", "crtQ","crtM","crtN","","","","","","","","")
And then launched ggplot with the specificities I wanted.
ggplot(BGC1, aes(xmin = start1, xmax = end1,y='', fill = Genes, forward = orientation1, label=C1))
geom_gene_arrow() geom_gene_label(y=1.03, size=20)
facet_wrap(~ Molecule, scales = "free", ncol = 1) scale_fill_brewer(palette = "Set2")
labs(x='locus (bp)', y='') theme(axis.title.x=element_text()) theme_minimal()
So, questions are:
- first, as you may notice the title of the graph is basically the name of the column Molecule in the dataframe, not a title per se. How could I edit its size and position?
- second, the title and x-axis are very far away from the genes. I'd like to compact everything a bit more to reduce that empty space (I learned how to move the 'locus (bp)' text but not the bottom of the graph)
- third, is there a way to set the 'unknown' gene colours to white or transparent? I saw it in the palette Set2 and I liked it.
CodePudding user response:
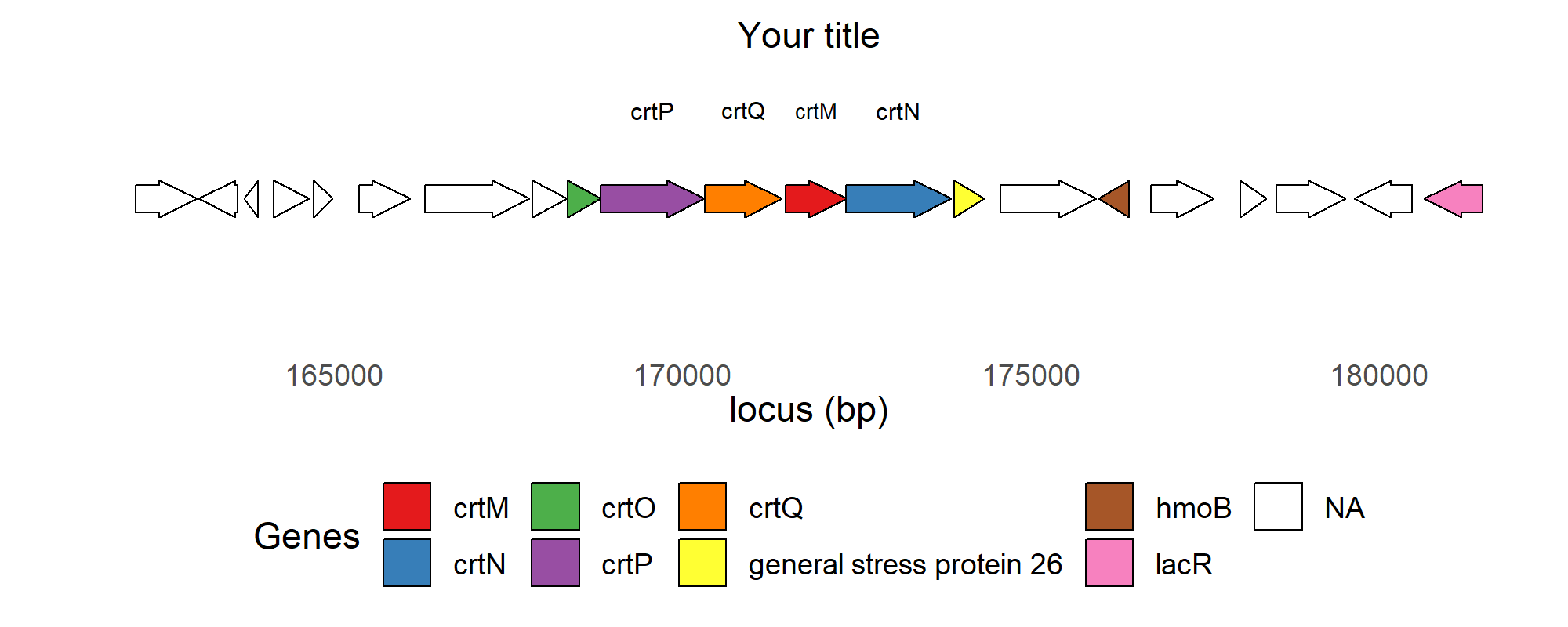
Here is a possible solution to your problems.
library(gggenes)
library(ggplot2)
Molecule <- rep("Staphyloxanthin synthesis-related BGC", 21)
start1 <- c(162155,163052,163705,164135,164702,165358,166305,167847,
168352,168826,170323,171480,172352,173908,174568,175973,
176730,178006,178528,179646,180648)
end1 <- c(163024,163609,163911,164641,164977,166086,167804,168347,
168825,170316,171426,172355,173857,174333,175941,176410,
177626,178377,179523,180476,181487)
Genes <- c("unknown","unknown","unknown","unknown","unknown","unknown",
"unknown","unknown","crtO","crtP","crtQ","crtM","crtN",
"general stress protein 26","unknown","hmoB","unknown",
"unknown","unknown","unknown","lacR")
strand1 <- c("forward","reverse","reverse","forward","forward","forward",
"forward","forward","forward","forward","forward","forward",
"forward","reverse","forward","forward",
"forward","reverse","reverse","forward","reverse")
orientation1 <- c(1,-1,-1,1,1,1,1,1,1,1,1,1,1,1,1,-1,1,1,1,-1,-1)
C1 <- c("","","","","","","","","crtO","crtP", "crtQ","crtM","crtN","",
"","","","","","","")
BGC1 <- data.frame(Molecule, start1, end1, Genes, strand1, orientation1)
BGC1$Genes[BGC1$Genes=="unknown"] <- NA
BGC1$Genes <- factor(BGC1$Genes)
BGC1$y <- 1
dev.new(width=8, height=3)
p <- ggplot(BGC1, aes(xmin = start1, xmax = end1, y=y,
fill = Genes, forward = orientation1, label=C1))
geom_gene_arrow()
geom_gene_label(y=1.03, size=20)
scale_fill_brewer(palette = "Set1", na.value=NA)
labs(x='locus (bp)', y='', title='Your title')
theme_minimal() % replace% theme(axis.title.x=element_text(),
axis.ticks.y = element_blank(),
axis.text.y=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.title = element_text(hjust = 0.5),
legend.direction="horizontal",
legend.position="bottom")
print(p)
If you need to save your plot in a file:
png(file="Figure.png", width=2000, height=800, res=300)
print(p)
dev.off()