I have a dataframe with ~40k lines and 52 columns. It describes locations of interest, with some overlap.
I am trying to filter out locations that overlap, so that I only keep the last entry.
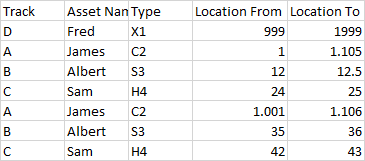
An example dataset is:
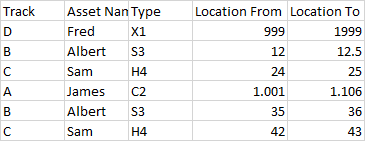
The expected output is:
The first Track A entry is to be excluded.
My code is below:
df_length = len(df.axes[0])
duplicate_flag = False
i = 0
trim_value = 0.005
data = []
for j in range(df_length - 1):
for k in range(df_length - 1):
if k >= i:
if(df.at[i, 'Track'] == df.at[k 1, 'Track'] and
df.at[i, 'Asset Name'] == df.at[k 1, 'Asset Name'] and
df.at[i, 'Type'] == df.at[k 1, 'Type'] and
df.at[i, 'Location From'] >= (df.at[k 1, 'Location From'] - trim_value) and
df.at[i, 'Location From'] <= (df.at[k 1, 'Location From'] trim_value) and
df.at[i, 'Location To'] >= (df.at[k 1, 'Location To'] - trim_value) and
df.at[i, 'Location To'] <= (df.at[k 1, 'Location To'] trim_value)):
duplicate_flag = True
if duplicate_flag == False:
data.append(df.iloc[i])
i = 1
duplicate_flag = False
data.append(df.iloc[-1])
df2 = pd.DataFrame(data)
For each row of the dataframe, I compare it to each row below it. If the Track, Asset Name and Type all equal, and the Location From and To values are or - the trim value, I set a duplicate flag as true.
After comparing to the rows below, if there was no duplicate found, I append that row of the original dataframe to a list, which is later converted into another dataframe.
I then increment the counter i, and reset the duplicate_flag variable to False.
My issue is this takes an extremely long time to run even with only 40k lines. I learnt that appending to a list and not to a dataframe is much more efficient, but I'm unsure what else is causing the bottleneck.
My intuition is suggesting it is the conditional if statements, though unsure what would be an improvement.
Any suggestions appreciated.
Thanks.
CodePudding user response:
dict = {'Track': ['D','A','B','C','A','B','C'],
'Asset Name':['Fred','James','Albert','Sam','James','Albert','Sam'],
'Type':['X1','C2','S3','H4','C2','S3','H4'],
'Location From':[999,1,12,24,1.001,35,42],
'Location To':[1999,1.105,12.5,25,1.106,36,43]}
df = pd.DataFrame(data=dict)
Tolerance=0.02
series = df
series = df.iloc[:,3:5].mul(1/Tolerance).round()
series = series.duplicated(keep='last')
Duplicate_Index = series[series].index
df = df.drop(Duplicate_Index)
print(df)
CodePudding user response:
I ended up experimenting with a list of dictionaries instead and the performance for this task in magnitudes quicker.
dict_list = df.to_dict('records')
dict_length = len(dict_list)
duplicate_flag = False
i = 0
trim_value = 0.005
data = []
for j in range(dict_length - 1):
for k in range(dict_length - 1):
if k >= i:
if(dict_list[i]['Running Date'] == dict_list[k 1]['Running Date'] and
dict_list[i]['Category'] == dict_list[k 1]['Category'] and
dict_list[i]['Location From'] >= (dict_list[k 1]['Location From'] - trim_value) and
dict_list[i]['Location From'] <= (dict_list[k 1]['Location From'] trim_value) and
dict_list[i]['Location To'] >= (dict_list[k 1]['Location To'] - trim_value) and
dict_list[i]['Location To'] <= (dict_list[k 1]['Location To'] trim_value)):
duplicate_flag = True
if duplicate_flag == False:
data.append(dict_list[i])
i = 1
duplicate_flag = False
data.append(dict_list[-1])
df2 = pd.DataFrame(data)