I've the a data frame containing different items (and it's cost) and also it's subsequent groupings. I would like to run an Anova and/or T-Test for each item based on their groupings to see if their mean differs. Anybody knows how to do this in R?
A sample of the dataframe is as follow:
| Item | Cost | Grouping |
|---|---|---|
| Book A | 7 | A |
| Book A | 9 | B |
| Book A | 6 | A |
| Book A | 7 | B |
| Book B | 4 | A |
| Book B | 6 | B |
| Book B | 5 | A |
| Book B | 3 | C |
| Book C | 5 | C |
| Book C | 4 | A |
| Book C | 7 | C |
| Book C | 2 | B |
| Book C | 2 | B |
| Book D | 4 | A |
| Book D | 2 | C |
| Book D | 9 | C |
| Book D | 4 | A |
The output should be a simple table (or any similar table) as follows
| Item | P-Value (from ANOVA/t-test) (H0: Mean same for all groupings) |
|---|---|
| Book A | xxx |
| Book B | xxx |
| Book C | xxx |
| Book D | xxx |
Thanks in advance!
CodePudding user response:
Instead of dealing with multiple ANOVA, t-tests and worrisome (and potentially questionable) p-values, I would fit a single generalised linear mixed-effect model with group as a random effect. This is easy to do in a fully Bayesian way using rstanarm, which gives full posterior distributions for the means of every item. Instead of worrying about the suitability & interpretability of (multiple) hypothesis tests, we can then compare posterior distributions for the means directly.
library(rstanarm)
model <- stan_glmer(cost ~ 0 item (1 | group), data = df)
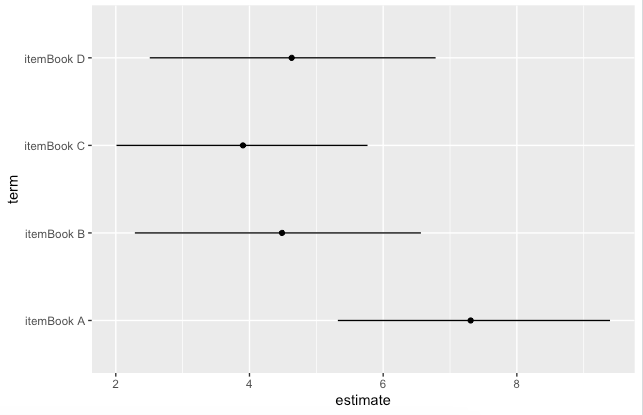
We can summarise the mean posterior distributions by showing the posterior median and 90% posterior uncertainty intervals per item.
library(broom.mixed)
tidy(model, conf.int = TRUE) %>%
ggplot(aes(y = term))
geom_point(aes(x = estimate))
geom_linerange(aes(xmin = conf.low, xmax = conf.high))
Or as a table
tidy(mode, conf.int = TRUE)
## A tibble: 4 × 5
# term estimate std.error conf.low conf.high
# <chr> <dbl> <dbl> <dbl> <dbl>
#1 itemBook A 7.28 1.17 5.09 9.40
#2 itemBook B 4.44 1.16 2.27 6.45
#3 itemBook C 3.88 1.05 1.89 5.75
#4 itemBook D 4.63 1.21 2.41 6.71
Here,
estimateis the posterior median,std.erroris the posterior MAD, andconf.lowandconf.highare the lower and upper bounds of the 90% posterior uncertainty interval.
CodePudding user response:
You could use anova_test from the rstatix package like this:
df <- data.frame(Item = c("Book A", "Book A", "Book A", "Book A", "Book B", "Book B", "Book B", "Book B"),
Cost = c(7,9,6,7,4,6,5,3),
Grouping = c("A", "B", "A", "B", "A", "B", "A", "C"))
library(dplyr)
library(rstatix)
df %>%
group_by(Item) %>%
anova_test(Cost ~ Grouping)
#> Coefficient covariances computed by hccm()
#> Coefficient covariances computed by hccm()
#> # A tibble: 2 × 8
#> Item Effect DFn DFd F p `p<.05` ges
#> * <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 Book A Grouping 1 2 1.8 0.312 "" 0.474
#> 2 Book B Grouping 2 1 4.5 0.316 "" 0.9
Created on 2022-07-10 by the reprex package (v2.0.1)