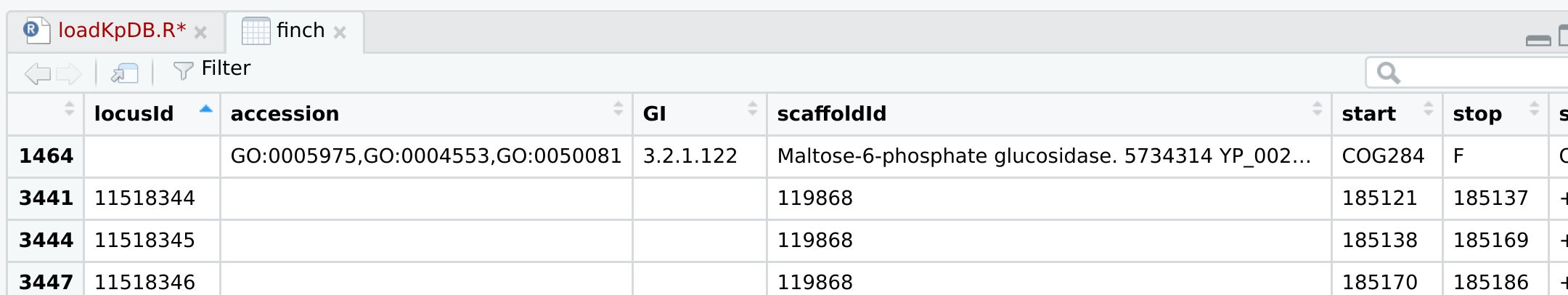
I am trying to load data into R, but some row does not work well. I got this issue a lot of time, but when I load them in excel, it works well. Please help me if you know the reason. Thank you very much!
library(RCurl)
URL <- "http://www.microbesonline.org/cgi-bin/genomeInfo.cgi?tId=507522;export=tab"
x <- getURL(URL, ssl.verifypeer = FALSE)
finch <- read.table(file = textConnection(x), header = 1, sep = "\t", fill = TRUE )
finch <- as.data.frame(finch)
CodePudding user response:
Use read.delim, it's a wrapper designed for tab delimited tables.
finch <- read.delim(textConnection(x))
It can deal with the problematic row:
> finch[1464, ]
locusId accession GI scaffoldId start stop strand sysName name
1464 5733093 YP_002237317.1 206579759 119869 1482145 1482804 KPK_1464 yfbT
desc COG COGFun COGDesc
1464 sugar-phosphatase, YfbT (RefSeq) COG637 R Predicted phosphatase/phosphohexomutase
TIGRFam TIGRRoles
1464 TIGR01509 HAD hydrolase, family IA, variant 3 Unknown function:Enzymes of unknown specificity
GO EC ECDesc
1464 GO:0008152,GO:0003824,GO:0050308 3.1.3.23 Sugar-phosphatase.