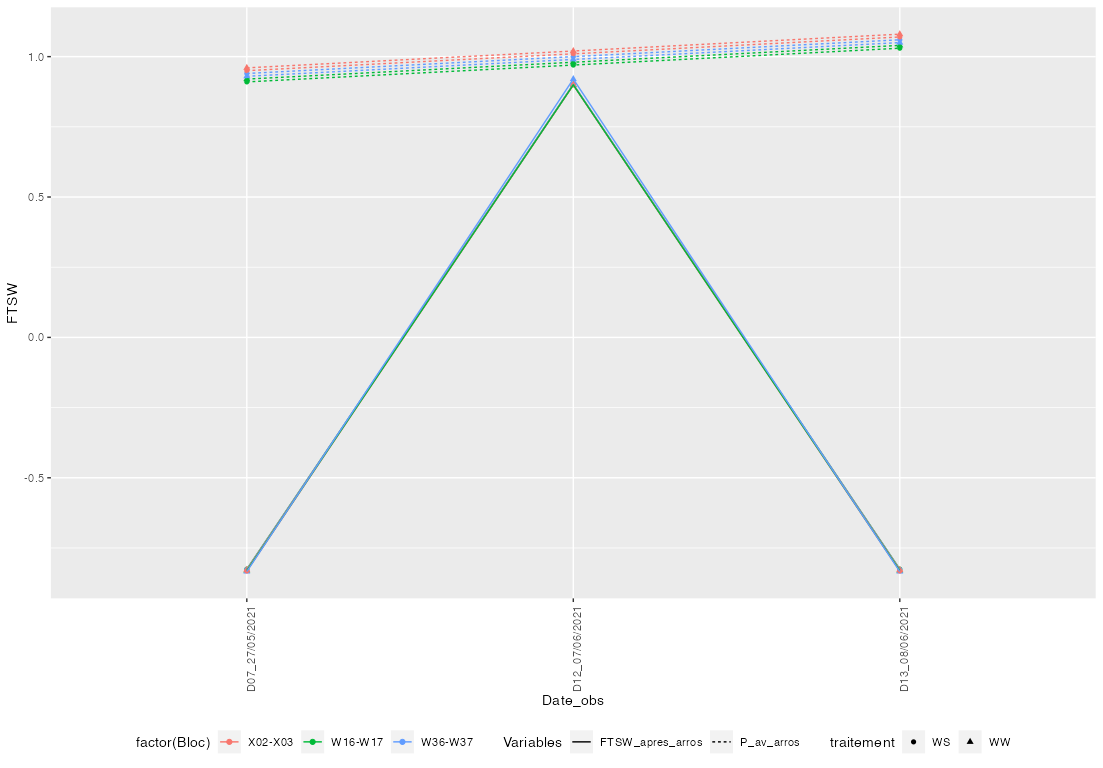
I want to draw two variables in a same figure, and for some details I want to distinguish them by the legend.
Please see the original data below:
df<-structure(list(Bloc = c(7, 7, 8, 8, 5, 5, 7, 7, 8, 8, 5, 5, 7,
7, 8, 8, 5, 5), Pos_heliaphen = c("W16", "W17", "W36", "W37",
"X02", "X03", "W16", "W17", "W36", "W37", "X02", "X03", "W16",
"W17", "W36", "W37", "X02", "X03"), traitement = c("WS", "WW",
"WW", "WS", "WS", "WW", "WS", "WW", "WW", "WS", "WS", "WW", "WS",
"WW", "WW", "WS", "WS", "WW"), Variete = c("Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas"), Date_obs = c("D07_27/05/2021",
"D07_27/05/2021", "D07_27/05/2021", "D07_27/05/2021", "D07_27/05/2021",
"D07_27/05/2021", "D12_07/06/2021", "D12_07/06/2021", "D12_07/06/2021",
"D12_07/06/2021", "D12_07/06/2021", "D12_07/06/2021", "D13_08/06/2021",
"D13_08/06/2021", "D13_08/06/2021", "D13_08/06/2021", "D13_08/06/2021",
"D13_08/06/2021"), P_av_arros = c(0.91, 0.92, 0.93, 0.94, 0.95,
0.96, 0.97, 0.98, 0.99, 1, 1.01, 1.02, 1.03, 1.04, 1.05, 1.06,
1.07, 1.08), FTSW_apres_arros = c(-0.82561716279966, -0.83052784248231,
-0.833812425397989, -0.826677385087996, -0.831991201718322, -0.827650244364889,
0.900083501645054, 0.899646933005172, 0.920126486265779, 0.901668054319428,
0.899616920453791, 0.899570142896103, -0.82561716279966, -0.83052784248231,
-0.833812425397989, -0.826677385087996, -0.831991201718322, -0.827650244364889
)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-18L))
I've already tried with the code:
library(ggplot2)
library(dplyr)
labels <- df %>%
select(Bloc, Pos_heliaphen) %>%
distinct(Bloc, Pos_heliaphen) %>%
group_by(Bloc) %>%
summarise(Pos_heliaphen = paste(Pos_heliaphen, collapse = "-")) %>%
tibble::deframe()
df %>%
select(1, 2, 3, 4, 5, 6, 7) %>%
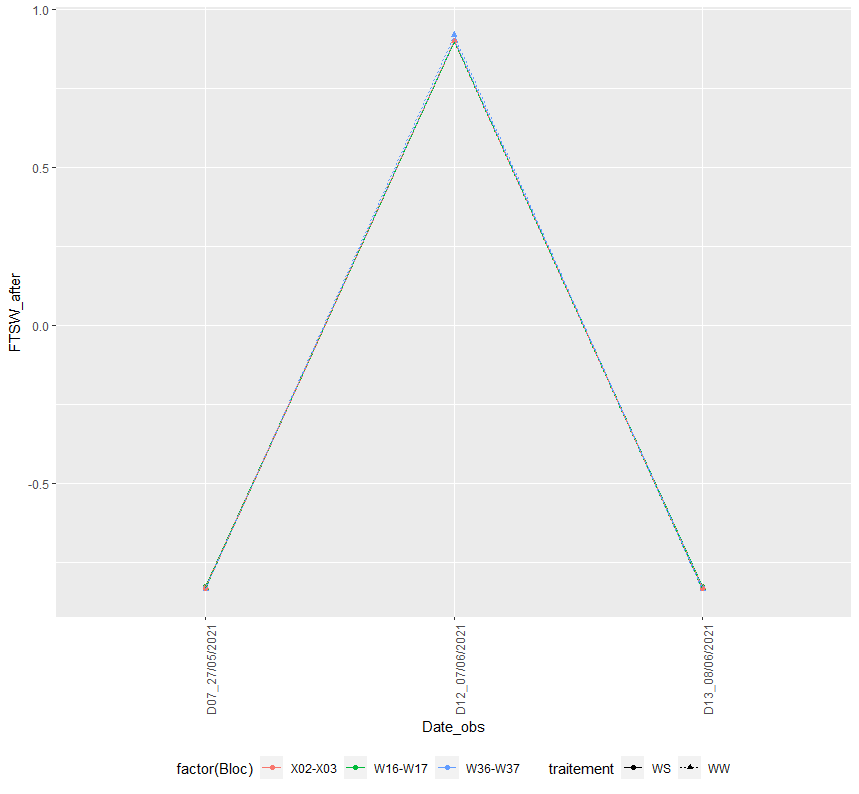
ggplot(aes(Date_obs, FTSW_apres_arros, colour = factor(Bloc), shape = traitement, linetype = traitement, group = interaction(Bloc, traitement)))
geom_point()
geom_line()
scale_color_discrete(labels = labels, guide = guide_legend(order = 1))
labs(y = expression(paste("FTSW")))
theme(legend.position = "bottom",
axis.text.x = element_text(angle = 90, hjust = 1))
And I gained the figure below:
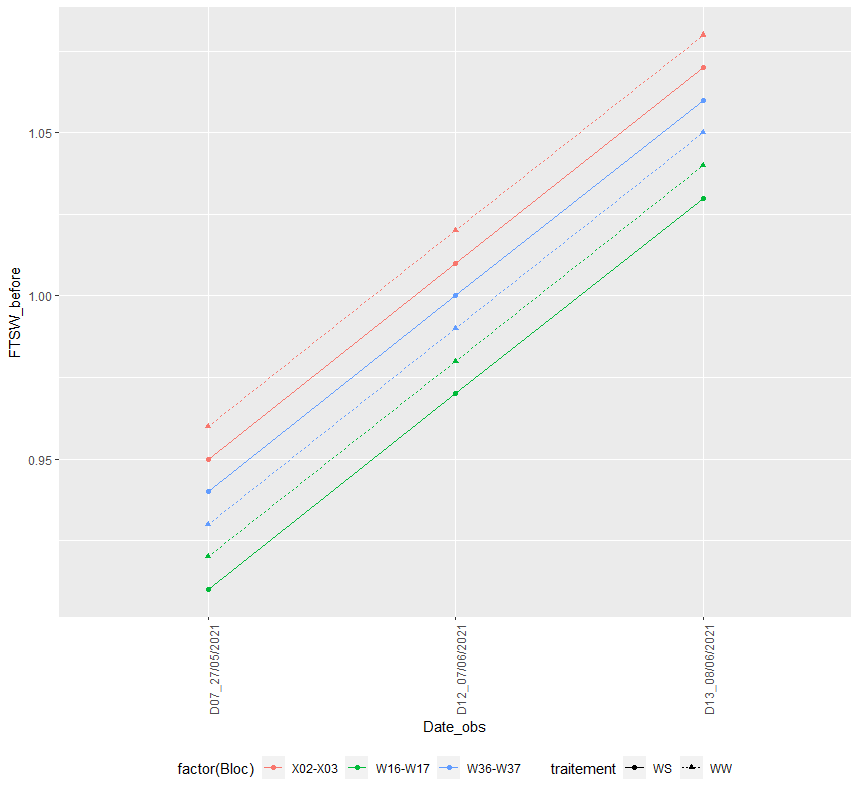
I can also the figure below if I change FTSW_apres_arros to P_av_arros:
Now I want to put the two variables FTSW_apres_arros and P_av_arrosin the same figure.
Below is the legend that I hope I can have:
I'm not sure if I explain my question clearly. Thanks in advance if anyone help me!
CodePudding user response:
One option to achieve your desired result would be to reshape your data to long before passing it to ggplot. This way you get a new Variables and a value column. The Variables column could be mapped on linetype while the value column is mapped on y. Additionally you have to add Variables to the interaction mapped on group. .
library(ggplot2)
library(dplyr)
library(tidyr)
df %>%
select(1, 2, 3, 4, 5, 6, 7) %>%
tidyr::pivot_longer(c(P_av_arros, FTSW_apres_arros), names_to = "Variables") %>%
ggplot(aes(Date_obs, value, colour = factor(Bloc), shape = traitement,
linetype = Variables, group = interaction(Bloc, traitement, Variables)))
geom_point()
geom_line()
scale_color_discrete(labels = labels, guide = guide_legend(order = 1))
labs(y = expression(paste("FTSW")))
theme(legend.position = "bottom",
axis.text.x = element_text(angle = 90, hjust = 1))
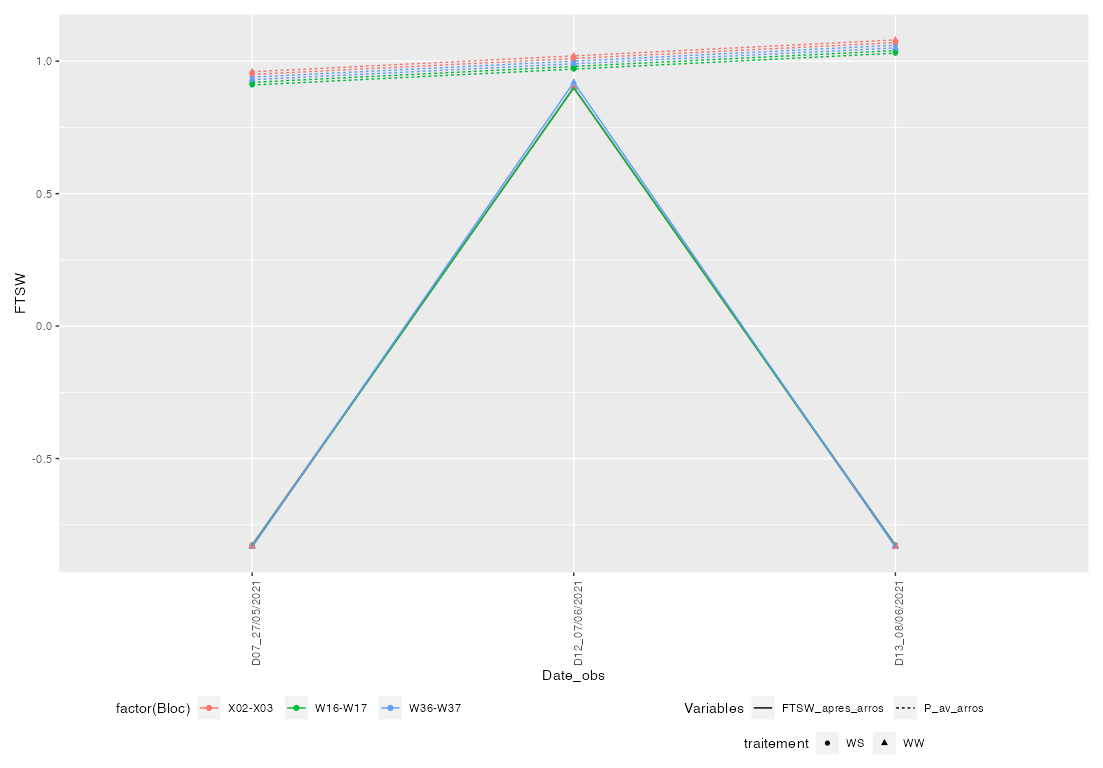
EDIT Unfortunately there is AFAIK no easy way to put legends in multiple rows and columns at the same time, i.e. using theme(legend.box.just = ...) you could have them all in one row (= "horizontal") or in one column (= "vertical").
One possible option would be to use cowplot::get_legend to extract the legends from your plot and then use patchwork to get closer to your desired result. However, when doing so the single legends will be centered and I'm afraid aligning them nicely requires a lot of effort.
library(ggplot2)
library(dplyr)
library(tidyr)
p <- df %>%
select(1, 2, 3, 4, 5, 6, 7) %>%
tidyr::pivot_longer(c(P_av_arros, FTSW_apres_arros), names_to = "Variables") %>%
ggplot(aes(Date_obs, value, colour = factor(Bloc), shape = traitement,
linetype = Variables, group = interaction(Bloc, traitement, Variables)))
geom_point()
geom_line()
scale_color_discrete(labels = labels, guide = guide_legend(order = 1))
labs(y = expression(paste("FTSW")))
theme(legend.position = "bottom",
axis.text.x = element_text(angle = 90, hjust = 1))
library(cowplot)
library(patchwork)
legends <- get_legend(p)
p1 <- p guides(color = "none", linetype = "none", shape = "none")
design <- "
AB
#C
"
p1 / (wrap_plots(legends$grobs[1:3], design = design)) plot_layout(heights = c(10, 1))