lets say I have the following data frame:
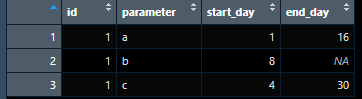
dt <- data.frame(id= c(1),
parameter= c("a","b","c"),
start_day = c(1,8,4),
end_day = c(16,NA,30))
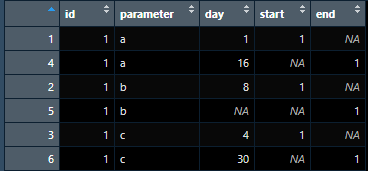
I need to combine start_day and end_day columns (lets call the new column as day) such that I reserve all the other columns. Also I need to create another column that indicates if each row is showing start_day or end_day. To clarify, I am looking to create the following data frame
I am creating the above data frame using the following code:
dt1 <- subset(dt, select = -c(end_day))
dt1 <- dt1 %>% rename(day = start_day)
dt1$start <- 1
dt2 <- subset(dt, select = -c(start_day))
dt2 <- dt2 %>% rename(day = end_day)
dt2$end <- 1
dt <- bind_rows(dt1, dt2)
dt <- dt[order(dt$id, dt$parameter),]
Although my code works, but I am not happy with my solution. I am certain that there is a better and cleaner way to do that. I would appreciate any input on better alternatives of tackling this problem.
CodePudding user response:
(tidyr::pivot_longer(dt, cols = c(start_day, end_day), values_to = "day")
|> dplyr::mutate(start = ifelse(name == "start_day", 1, NA),
end = ifelse(name == "end_day", 1, NA))
)
Result:
# A tibble: 6 × 6
id parameter name day start end
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 1 a start_day 1 1 NA
2 1 a end_day 16 NA 1
3 1 b start_day 8 1 NA
4 1 b end_day NA NA 1
5 1 c start_day 4 1 NA
6 1 c end_day 30 NA 1
You could get rid of the name column, but maybe it would be more useful than your new start/end columns?
CodePudding user response:
using base R (faster than data.table up to ~300 rows; faster than tidyr up to ~1k rows) :
cbind(dt[1:2], day = c(dt$start_day,dt$end_day)) |>
(\(x) x[order(x$id, x$parameter),])() |>
(`[[<-`)("start", value = c(1, NA)) |>
(`[[<-`)("end", value = c(NA, 1))
id parameter day start end
1 1 a 1 1 NA
4 1 a 16 NA 1
2 1 b 8 1 NA
5 1 b NA NA 1
3 1 c 4 1 NA
6 1 c 30 NA 1
using the data.table package (faster than tidyr up to ~500k rows) :
dt <- as.data.table(dt)
dt[,.(day = c(start_day, end_day),
start = rep(c(1, NA), .N),
end = rep(c(NA, 1), .N)),
by = .(id, parameter)]
id parameter day start end
1: 1 a 1 1 NA
2: 1 a 16 NA 1
3: 1 b 8 1 NA
4: 1 b NA NA 1
5: 1 c 4 1 NA
6: 1 c 30 NA 1