I have the following data:
| Category | albumin | protein | cholesterol | urea |
|---|---|---|---|---|
| placebo | 24,04 | 61,45 | 63,09 | 19,04 |
| placebo | 25,86 | 57,63 | 111,04 | 14,30 |
| placebo | 28,18 | 64,18 | 108 | 18,68 |
| test | 23,77 | 61,45 | 70,59 | 17,43 |
| test | 26,22 | 57,54 | 123,45 | 13,68 |
| test | 27,90 | 65,36 | 106,22 | 18,63 |
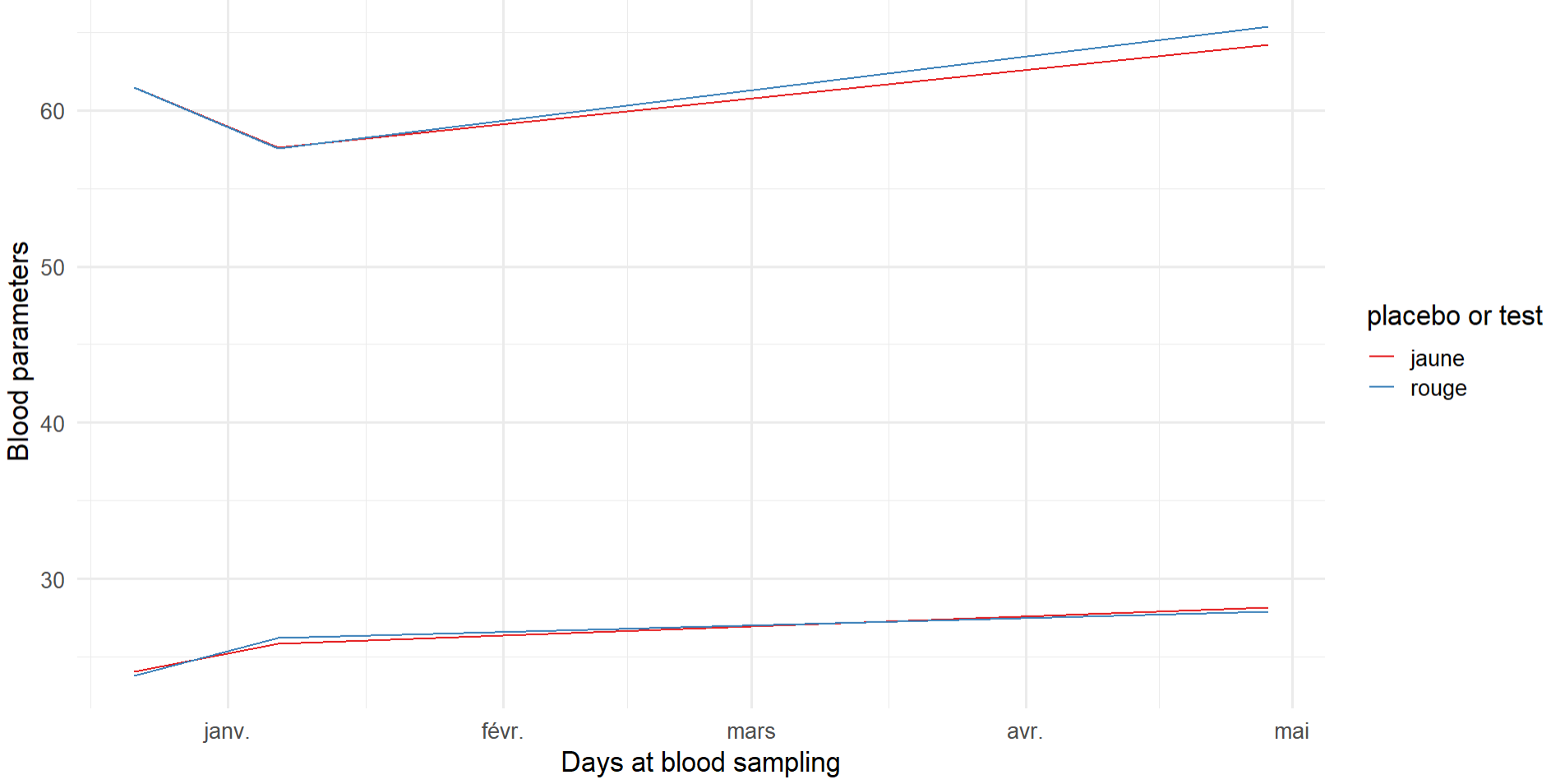
And I would like to plot the different blood parameters on the same plot with 2 different colors for the category placebo or test. Here is the code and what I have:
plot <- ggplot()
geom_line(data = file, aes(x = date, y = albumin, color = category))
geom_line( data = file, aes(x = date, y = protein, color = category))
scale_color_brewer(palette = "Set1")
theme_minimal(base_size = 16)
plot xlab("Days at blood sampling") ylab("Blood parameters")
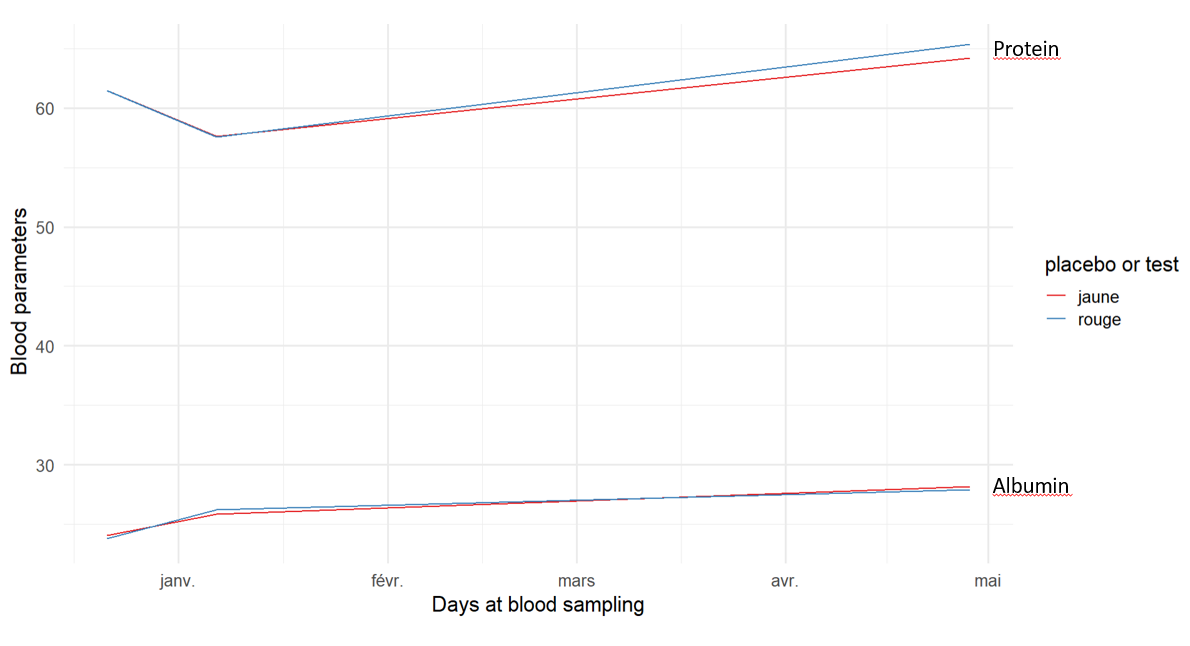
My issue is to add a label with the name of each blood parameter next to each line, a bit like this:
How can I do this?
CodePudding user response:
One option to achieve your desired result would be "the secondary axis trick" which adds the labels as breaks and labels of a secondary y axis. To this end I first create a dataframe containing the values for the breaks (for which I chose the mean value) and the labels:
library(dplyr, warn = FALSE)
library(tidyr)
library(ggplot2)
breaks <- file |>
filter(date == max(date)) |>
summarise(across(c(albumin, protein), mean)) |>
tidyr::pivot_longer(cols = everything())
ggplot(file,aes(x = date))
geom_line(aes(y = albumin, color = Category))
geom_line( aes(y = protein, color = Category))
scale_color_brewer(palette = "Set1")
scale_y_continuous(sec.axis = dup_axis(breaks = breaks$value, labels = stringr::str_to_title(breaks$name)), name = NULL)
theme_minimal(base_size = 16)
labs(x = "Days at blood sampling", y = "Blood parameters")
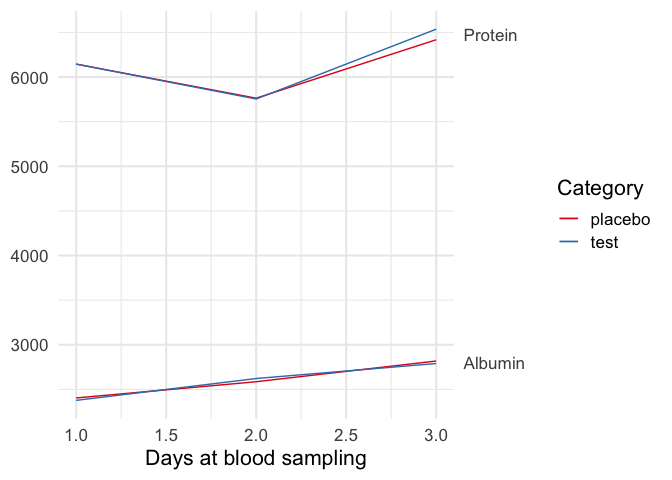
DATA
file <- data.frame(
stringsAsFactors = FALSE,
Category = c("placebo", "placebo", "placebo", "test", "test", "test"),
albumin = c(2404, 2586, 2818, 2377, 2622, 2790),
protein = c(6145, 5763, 6418, 6145, 5754, 6536),
cholesterol = c(6309, 11104, 108, 7059, 12345, 10622),
urea = c(1904, 1430, 1868, 1743, 1368, 1863)
)
file$date <- rep(1:3, 2)