Problem:
For each row of a DataFrame, I want to find the nearest prior row where the 'Datetime' value is at least 20 seconds before the current 'Datetime' value.
For example: if the previous 'Datetime' (at index i-1) is at least 20s earlier than the current one - it will be chosen. Otherwise (e.g. only 5 seconds earlier), move to i-2 and see if it is at least 20s earlier. Repeat until the condition is met, or no such row has been found.
The expected result is a concatenation of the original df and the rows that were found. When no matching row at or more than 20 s before the current Datetime has been found, then the new columns are null (NaT or NaN, depending on the type).
Example data
df = pd.DataFrame({
'Datetime': pd.to_datetime([
f'2016-05-15 08:{M_S} 06:00'
for M_S in ['36:21', '36:41', '36:50', '37:10', '37:19', '37:39']]),
'A': [21, 43, 54, 2, 54, 67],
'B': [3, 3, 45, 23, 8, 6],
})
Example result:
>>> res
Datetime A B Datetime_nearest A_nearest B_nearest
0 2016-05-15 08:36:21 06:00 21 3 NaT NaN NaN
1 2016-05-15 08:36:41 06:00 43 3 2016-05-15 08:36:21 06:00 21.0 3.0
2 2016-05-15 08:36:50 06:00 54 45 2016-05-15 08:36:21 06:00 21.0 3.0
3 2016-05-15 08:37:10 06:00 2 23 2016-05-15 08:36:50 06:00 54.0 45.0
4 2016-05-15 08:37:19 06:00 54 8 2016-05-15 08:36:50 06:00 54.0 45.0
5 2016-05-15 08:37:39 06:00 67 6 2016-05-15 08:37:19 06:00 54.0 8.0
The last three columns are the newly created columns, and the first three columns are the original dataset.
CodePudding user response:
Two vectorized solutions
Note: we assume that the rows are sorted by Datetime. If that is not the case, then sort them first (O[n log n]).
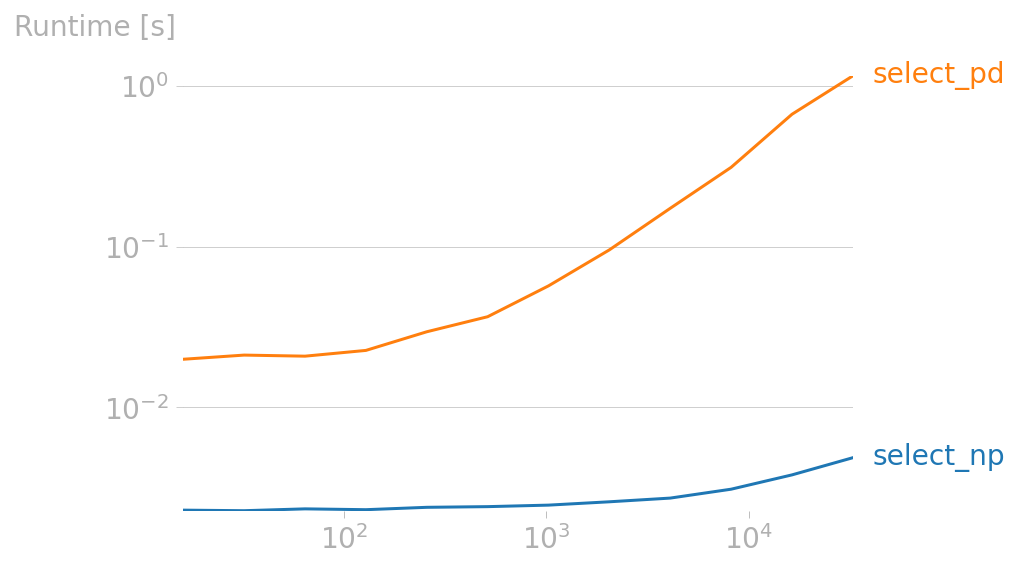
For 10,000 rows:
- 3.3 ms, using Numpy's
searchsorted. - 401 ms, using a
rollingwindow of 20s, left-open.
1. Using np.searchsorted
We use np.searchsorted to find in one call the index of all previous rows. E.g., for the OP's data, these indices are:
import numpy as np
s = df['Datetime']
z = np.searchsorted(s, s - (pd.Timedelta(min_dt) - pd.Timedelta('1ns'))) - 1
>>> z
array([-1, 0, 0, 2, 2, 4])
I.e.: z[0] == -1: no matching row; z[1] == 0: row 0 (08:36:21) is the nearest that is 20s or more before row 1 (08:36:41). z[2] == 0: row 0 is the nearest match for row 2 (row 1 is too close). Etc.
Why subtracting 1? We use np.searchsorted to select the first row in the exclusion zone (i.e., too close); then we subtract 1 to get the correct row (the first one at least 20s before).
Why - 1ns? This is to make the search window left-open. A row at exactly 20s before the current one will not be in the exclusion zone, and thus will end up being the one selected as the match.
We then use z to select the matching rows (or nulls) and concatenate into the result. Putting it all in a function:
def select_np(df, min_dt='20s'):
newcols = [f'{k}_nearest' for k in df.columns]
s = df['Datetime']
z = np.searchsorted(s, s - (pd.Timedelta(min_dt) - pd.Timedelta('1ns'))) - 1
return pd.concat([
df,
df.iloc[z].set_axis(newcols, axis=1).reset_index(drop=True).where(pd.Series(z >= 0))
], axis=1)
On the OP's example
>>> select_np(df[['Datetime', 'A', 'B']])
Datetime A B Datetime_nearest A_nearest B_nearest
0 2016-05-15 08:36:21 06:00 21 3 NaT NaN NaN
1 2016-05-15 08:36:41 06:00 43 3 2016-05-15 08:36:21 06:00 21.0 3.0
2 2016-05-15 08:36:50 06:00 54 45 2016-05-15 08:36:21 06:00 21.0 3.0
3 2016-05-15 08:37:10 06:00 2 23 2016-05-15 08:36:50 06:00 54.0 45.0
4 2016-05-15 08:37:19 06:00 54 8 2016-05-15 08:36:50 06:00 54.0 45.0
5 2016-05-15 08:37:39 06:00 67 6 2016-05-15 08:37:19 06:00 54.0 8.0
2. Using a rolling window (pure Pandas)
This was our original solution and uses pandas rolling with a Timedelta(20s) window size, left-open. It is still more optimized than a naive (O[n^2]) search, but is roughly 100x slower than select_np(), as pandas uses explicit loops in Python to find the window bounds for .rolling(): see get_window_bounds(). There is also some overhead due to having to make sub-frames, applying a function or aggregate, etc.
def select_pd(df, min_dt='20s'):
newcols = [f'{k}_nearest' for k in df.columns]
z = (
df.assign(rownum=range(len(df)))
.rolling(pd.Timedelta(min_dt), on='Datetime', closed='right')['rownum']
.apply(min).astype(int) - 1
)
return pd.concat([
df,
df.iloc[z].set_axis(newcols, axis=1).reset_index(drop=True).where(z >= 0)
], axis=1)
3. Testing
First, we write an arbitrary-size test data generator:
def gen(n):
return pd.DataFrame({
'Datetime': pd.Timestamp('2020') \
np.random.randint(0, 30, n).cumsum() * pd.Timedelta('1s'),
'A': np.random.randint(0, 100, n),
'B': np.random.randint(0, 100, n),
})
Example
np.random.seed(0)
tdf = gen(10)
>>> select_np(tdf)
Datetime A B Datetime_nearest A_nearest B_nearest
0 2020-01-01 00:00:12 21 87 NaT NaN NaN
1 2020-01-01 00:00:27 36 46 NaT NaN NaN
2 2020-01-01 00:00:48 87 88 2020-01-01 00:00:27 36.0 46.0
3 2020-01-01 00:00:48 70 81 2020-01-01 00:00:27 36.0 46.0
4 2020-01-01 00:00:51 88 37 2020-01-01 00:00:27 36.0 46.0
5 2020-01-01 00:01:18 88 25 2020-01-01 00:00:51 88.0 37.0
6 2020-01-01 00:01:21 12 77 2020-01-01 00:00:51 88.0 37.0
7 2020-01-01 00:01:28 58 72 2020-01-01 00:00:51 88.0 37.0
8 2020-01-01 00:01:37 65 9 2020-01-01 00:00:51 88.0 37.0
9 2020-01-01 00:01:56 39 20 2020-01-01 00:01:28 58.0 72.0 ```
**Speed**
```python
tdf = gen(10_000)
% timeit select_np(tdf)
3.31 ms ± 6.79 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
%timeit select_pd(df)
401 ms ± 1.66 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
>>> select_np(df).equals(select_pd(df))
True
Scale sweep
We can now compare speed over a range of sizes, using the excellent perfplot package:
import perfplot
perfplot.plot(
setup=gen,
kernels=[select_np, select_pd],
n_range=[2**k for k in range(4, 16)],
equality_check=lambda a, b: a.equals(b),
)
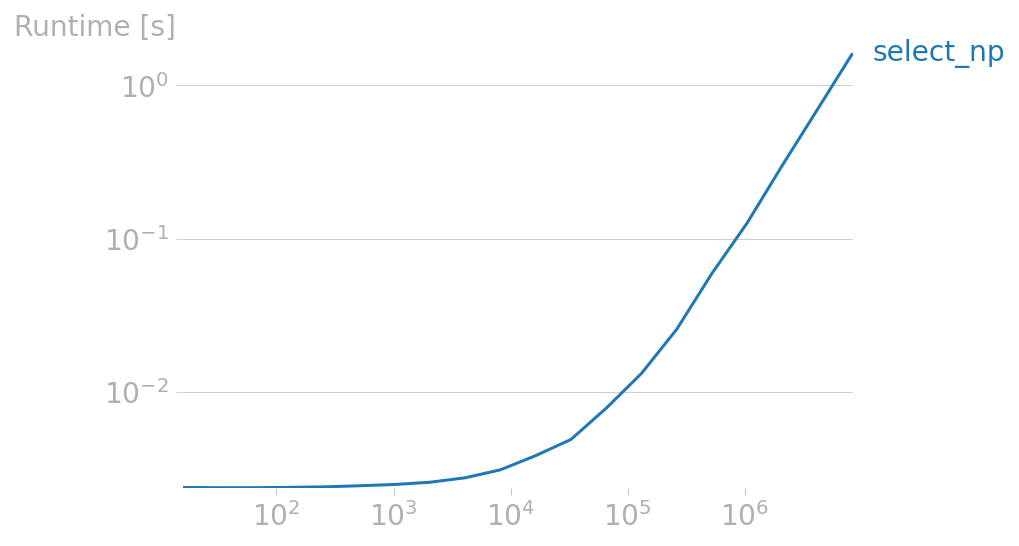
Focusing on select_np:
perfplot.plot(
setup=gen,
kernels=[select_np],
n_range=[2**k for k in range(4, 24)],
)
CodePudding user response:
The following solution is memory-efficient but it is not the fastest one (because it uses iteration over rows).
The fully vectorized version (that I could think of on my own) would be faster but it would use O(n^2) memory.
Example dataframe:
timestamps = [pd.Timestamp('2016-01-01 00:00:00'),
pd.Timestamp('2016-01-01 00:00:19'),
pd.Timestamp('2016-01-01 00:00:20'),
pd.Timestamp('2016-01-01 00:00:21'),
pd.Timestamp('2016-01-01 00:00:50')]
df = pd.DataFrame({'Datetime': timestamps,
'A': np.arange(10, 15),
'B': np.arange(20, 25)})
| Datetime | A | B | |
|---|---|---|---|
| 0 | 2016-01-01 00:00:00 | 10 | 20 |
| 1 | 2016-01-01 00:00:19 | 11 | 21 |
| 2 | 2016-01-01 00:00:20 | 12 | 22 |
| 3 | 2016-01-01 00:00:21 | 13 | 23 |
| 4 | 2016-01-01 00:00:50 | 14 | 24 |
Solution:
times = df['Datetime'].to_numpy() # it's convenient to have it as an `ndarray`
shifted_times = times - pd.Timedelta(20, unit='s')
usefulis a list of "useful" indices ofdf- i.e. where the appended values will NOT benan:
useful = np.nonzero(shifted_times >= times[0])[0]
# useful == [2, 3, 4]
- Truncate
shifted_timesfrom the beginning - to iterate through useful elements only:
if len(useful) == 0:
# all new columns will be `nan`s
first_i = 0 # this value will never actually be used
useful_shifted_times = np.array([], dtype=shifted_times.dtype)
else:
first_i = useful[0] # first_i == 2
useful_shifted_times = shifted_times[first_i : ]
Find the corresponding index positions of
dffor each "useful" value.(these index positions are essentially the indices of
timesthat are selected for each element ofuseful_shifted_times):
selected_indices = []
# Iterate through `useful_shifted_times` one by one:
# (`i` starts at `first_i`)
for i, shifted_time in enumerate(useful_shifted_times, first_i):
selected_index = np.nonzero(times[: i] <= shifted_time)[0][-1]
selected_indices.append(selected_index)
# selected_indices == [0, 0, 3]
- Selected rows:
df_nearest = df.iloc[selected_indices].add_suffix('_nearest')
| Datetime_nearest | A_nearest | B_nearest | |
|---|---|---|---|
| 0 | 2016-01-01 00:00:00 | 10 | 20 |
| 0 | 2016-01-01 00:00:00 | 10 | 20 |
| 3 | 2016-01-01 00:00:21 | 13 | 23 |
Replace indices of
df_nearestto match those of the corresponding rows ofdf.(basically, that is the last
len(selected_indices)indices):
df_nearest.index = df.index[len(df) - len(selected_indices) : ]
| Datetime_nearest | A_nearest | B_nearest | |
|---|---|---|---|
| 2 | 2016-01-01 00:00:00 | 10 | 20 |
| 3 | 2016-01-01 00:00:00 | 10 | 20 |
| 4 | 2016-01-01 00:00:21 | 13 | 23 |
- Append the selected rows to the original dataframe to get the final result:
new_df = df.join(df_nearest)
| Datetime | A | B | Datetime_nearest | A_nearest | B_nearest | |
|---|---|---|---|---|---|---|
| 0 | 2016-01-01 00:00:00 | 10 | 20 | NaT | nan | nan |
| 1 | 2016-01-01 00:00:19 | 11 | 21 | NaT | nan | nan |
| 2 | 2016-01-01 00:00:20 | 12 | 22 | 2016-01-01 00:00:00 | 10 | 20 |
| 3 | 2016-01-01 00:00:21 | 13 | 23 | 2016-01-01 00:00:00 | 10 | 20 |
| 4 | 2016-01-01 00:00:50 | 14 | 24 | 2016-01-01 00:00:21 | 13 | 23 |
Note: NaT stands for 'Not a Time'. It is the equivalent of nan for time values.
Note: it also works as expected even if all the last 'Datetime' - 20 sec is before the very first 'Datetime' --> all new columns will be nans.
