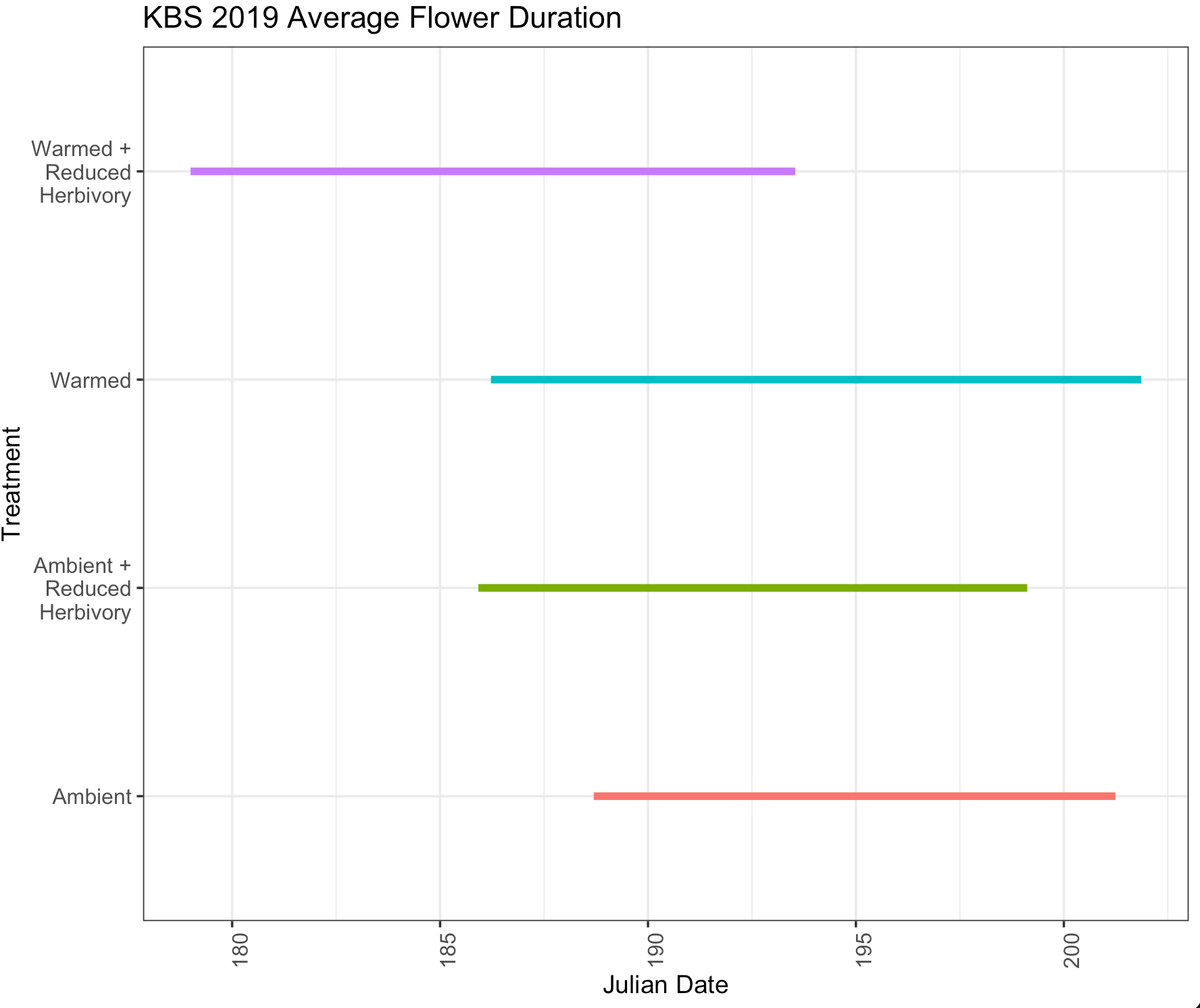
I want to make a horizontal line chart with date on the x axis and y axis is treatment with there being multiple lines for each year of data for average Julian date of first flower for each treatment. I don't know how to make this plot to include multiple years.
I want something like this (see image below) but there would be 2 lines for each treatment type to represent each year.
Code I used for the above plot:
ggplot(flwr_avg_duration_19)
geom_segment(aes(x = avg_first_flwr, xend = avg_last_flwr, y = treatment_key, yend = treatment_key, color= as.factor(treatment_key)), size = 2)
labs(title = "KBS 2019 Average Flower Duration", x = "Julian Date", y = "Treatment")
scale_y_discrete(labels=c("A0" = "Ambient",
"AI" = "Ambient \n Reduced\n Herbivory",
"W0"="Warmed",
"WI"="Warmed \n Reduced\n Herbivory"))
theme(legend.position = "none")
Here is a subset of my data:
dput(flwr_avg_duration_umbs[1:8,])
structure(list(site = c("umbs", "umbs", "umbs", "umbs", "umbs",
"umbs", "umbs", "umbs"), year = c(2016L, 2016L, 2016L, 2016L,
2017L, 2017L, 2017L, 2017L), state = c("ambient", "ambient",
"warmed", "warmed", "ambient", "ambient", "warmed", "warmed"),
treatment_key = c("A0", "AI", "W0", "WI", "A0", "AI", "W0",
"WI"), insecticide = c("no_insects", "insects", "no_insects",
"insects", "no_insects", "insects", "no_insects", "insects"
), year_factor = c(2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L), avg_first_flwr = c(157.5,
162.904761904762, 154.76, 160.090909090909, 160.678571428571,
161.269230769231, 159.848484848485, 158.695652173913), avg_last_flwr = c(182.384615384615,
186.761904761905, 186.64, 186.727272727273, 187.5, 188.423076923077,
186.939393939394, 187.608695652174)), row.names = 29:36, class = "data.frame")
CodePudding user response:
You need position_dodge to show the groups by year. Use the group aesthetic to ensure you are dodging by year. You will need to use geom_linerange instead of geom_segment, but that is the natural choice here anyway.
ggplot(flwr_avg_duration_umbs)
geom_linerange(aes(xmin = avg_first_flwr, xmax = avg_last_flwr,
y = treatment_key,
color = as.factor(treatment_key),
group = year), size = 2,
position = position_dodge(width = 0.5))
labs(title = "KBS 2019 Average Flower Duration",
x = "Julian Date", y = "Treatment")
scale_y_discrete(labels = c("A0" = "Ambient",
"AI" = "Ambient \n Reduced\n Herbivory",
"W0" = "Warmed",
"WI" = "Warmed \n Reduced\n Herbivory"))
theme_bw()
theme(legend.position = "none")