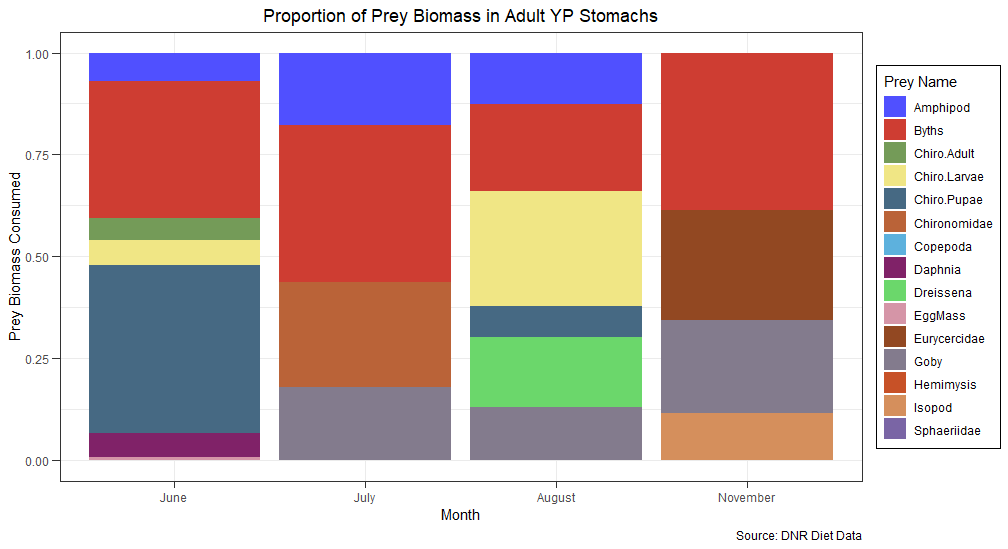
I would like to add labels on top of my bar plot (ggplot) to be able to visually see my groups easier.
Here is a condensed version of my data:
structure(list(prey_name = c("Empty", "Dreissena", "Amphipod",
"Chiro.Pupae", "Goby", "Byths", "Chiro.Larvae", "Byths", "Empty",
"Empty", "Byths", "Amphipod", "Eurycercidae", "Empty", "Chiro.Pupae",
"Empty", "Amphipod", "Goby", "Daphnia", "Chiro.Larvae", "Byths",
"Byths", "Byths", "Empty", "Sphaeriidae", "Empty", "Empty", "Chiro.Pupae",
"Amphipod", "Empty", "Byths", "Chiro.Pupae", "Isopod", "Byths",
"Byths", "Empty", "Chiro.Adult", "Hemimysis", "Chiro.Pupae",
"Byths", "Empty", "Empty", "Empty", "Amphipod", "Empty", "Empty",
"Empty", "Empty", "Goby", "Empty", "Byths", "Chiro.Larvae", "Amphipod",
"Empty", "Chironomidae", "Goby", "Amphipod", "Empty", "Byths",
"Empty", "Chiro.Larvae", "Chiro.Larvae", "Byths", "Empty", "Dreissena",
"Chironomidae", "Chiro.Pupae", "Empty", "Isopod", "Dreissena",
"Byths", "Copepoda", "Goby", "Empty", "Empty", "EggMass", "Empty",
"Empty", "Amphipod", "Empty", "Amphipod", "Byths", "Eurycercidae",
"Sphaeriidae", "Empty", "Chiro.Pupae", "Chiro.Pupae", "Empty",
"Amphipod", "Empty", "Byths", "Byths", "Byths", "Goby", "Empty",
"Empty", "Empty", "Empty", "Empty", "Empty"), id = c("708_1",
"824_29", "723_80", "604_2", "1031_2", "719_34", "824_39", "619_25",
"802_5", "822_2", "602_10", "803_2", "1024_33", "630_13", "619_27",
"701_2", "627_3", "701_6", "647_16", "824_31", "643_9", "807_13",
"601_6", "601_7", "616_6", "712_3", "639_4", "652_1", "604_2",
"701_7", "643_8", "627_2", "616_6", "712_6", "824_30", "643_3",
"614_3", "803_6", "614_1", "629_2", "712_5", "703_1", "712_4",
"622_5", "824_21", "822_5", "630_13", "801_10", "1031_1", "807_11",
"1004_18", "652_1", "719_32", "807_1", "716_1", "1031_1", "713_1",
"1003_26", "1010_1", "822_6", "824_28", "647_17", "601_5", "630_11",
"824_35", "723_80", "813_3", "708_2", "1024_33", "824_26", "813_3",
"627_2", "803_6", "801_15", "1030_38", "643_7", "617_3", "605_4",
"813_3", "643_1", "817_6", "630_10", "1024_44", "617_1", "701_5",
"616_3", "635_4", "807_2", "614_1", "801_6", "808_1", "642_7",
"643_2", "804_1", "701_3", "617_4", "635_2", "801_3", "619_20",
"1029_6"), CRN = c(708L, 824L, 723L, 604L, 1031L, 719L, 824L,
619L, 802L, 822L, 602L, 803L, 1024L, 630L, 619L, 701L, 627L,
701L, 647L, 824L, 643L, 807L, 601L, 601L, 616L, 712L, 639L, 652L,
604L, 701L, 643L, 627L, 616L, 712L, 824L, 643L, 614L, 803L, 614L,
629L, 712L, 703L, 712L, 622L, 824L, 822L, 630L, 801L, 1031L,
807L, 1004L, 652L, 719L, 807L, 716L, 1031L, 713L, 1003L, 1010L,
822L, 824L, 647L, 601L, 630L, 824L, 723L, 813L, 708L, 1024L,
824L, 813L, 627L, 803L, 801L, 1030L, 643L, 617L, 605L, 813L,
643L, 817L, 630L, 1024L, 617L, 701L, 616L, 635L, 807L, 614L,
801L, 808L, 642L, 643L, 804L, 701L, 617L, 635L, 801L, 619L, 1029L
), FID = c(1L, 29L, 80L, 2L, 2L, 34L, 39L, 25L, 5L, 2L, 10L,
2L, 33L, 13L, 27L, 2L, 3L, 6L, 16L, 31L, 9L, 13L, 6L, 7L, 6L,
3L, 4L, 1L, 2L, 7L, 8L, 2L, 6L, 6L, 30L, 3L, 3L, 6L, 1L, 2L,
5L, 1L, 4L, 5L, 21L, 5L, 13L, 10L, 1L, 11L, 18L, 1L, 32L, 1L,
1L, 1L, 1L, 26L, 1L, 6L, 28L, 17L, 5L, 11L, 35L, 80L, 3L, 2L,
33L, 26L, 3L, 2L, 6L, 15L, 38L, 7L, 3L, 4L, 3L, 1L, 6L, 10L,
44L, 1L, 5L, 3L, 4L, 2L, 1L, 6L, 1L, 7L, 2L, 1L, 3L, 4L, 2L,
3L, 20L, 6L), number = c(0L, 1L, 99L, 1216L, 5L, 239L, 4L, 30L,
0L, 0L, 20L, 40L, 931L, 0L, 1L, 0L, 173L, 1L, 363L, 9L, 608L,
68L, 251L, 0L, 1L, 0L, 0L, 32L, 1L, 0L, 256L, 50L, 5L, 194L,
8L, 0L, 74L, 6L, 255L, 8L, 0L, 0L, 0L, 3L, 0L, 0L, 0L, 0L, 3L,
0L, 333L, 2L, 3L, 0L, 23L, 3L, 1L, 0L, 107L, 0L, 5L, 3L, 20L,
0L, 1L, 67L, 1L, 0L, 17L, 1L, 68L, 8L, 1L, 0L, 0L, 3L, 0L, 0L,
1L, 0L, 32L, 213L, 1056L, 3L, 0L, 576L, 55L, 0L, 6L, 0L, 330L,
6L, 72L, 2L, 0L, 0L, 0L, 0L, 0L, 0L), MONTH = c(7L, 8L, 7L, 6L,
11L, 7L, 8L, 6L, 8L, 8L, 6L, 8L, 11L, 6L, 6L, 7L, 6L, 7L, 6L,
8L, 6L, 8L, 6L, 6L, 6L, 7L, 6L, 6L, 6L, 7L, 6L, 6L, 6L, 7L, 8L,
6L, 6L, 8L, 6L, 6L, 7L, 7L, 7L, 6L, 8L, 8L, 6L, 8L, 11L, 8L,
11L, 6L, 7L, 8L, 7L, 11L, 7L, 11L, 11L, 8L, 8L, 6L, 6L, 6L, 8L,
7L, 8L, 7L, 11L, 8L, 8L, 6L, 8L, 8L, 11L, 6L, 6L, 6L, 8L, 6L,
8L, 6L, 11L, 6L, 7L, 6L, 6L, 8L, 6L, 8L, 8L, 6L, 6L, 8L, 7L,
6L, 6L, 8L, 6L, 11L), adult = c("Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "Y"), month_name = structure(c(7L, 8L, 7L, 6L, 11L,
7L, 8L, 6L, 8L, 8L, 6L, 8L, 11L, 6L, 6L, 7L, 6L, 7L, 6L, 8L,
6L, 8L, 6L, 6L, 6L, 7L, 6L, 6L, 6L, 7L, 6L, 6L, 6L, 7L, 8L, 6L,
6L, 8L, 6L, 6L, 7L, 7L, 7L, 6L, 8L, 8L, 6L, 8L, 11L, 8L, 11L,
6L, 7L, 8L, 7L, 11L, 7L, 11L, 11L, 8L, 8L, 6L, 6L, 6L, 8L, 7L,
8L, 7L, 11L, 8L, 8L, 6L, 8L, 8L, 11L, 6L, 6L, 6L, 8L, 6L, 8L,
6L, 11L, 6L, 7L, 6L, 6L, 8L, 6L, 8L, 8L, 6L, 6L, 8L, 7L, 6L,
6L, 8L, 6L, 11L), levels = c("January", "February", "March",
"April", "May", "June", "July", "August", "September", "October",
"November", "December"), class = "factor"), BMASSprop = c(NaN,
0.0174639459714402, 0.09975608226961, 0.999816612301896, 0.2,
1, 0.99369653669798, 0.0119834315937307, NaN, NaN, 1, 1, 0.0318128167117478,
NaN, 0.991957112535848, NaN, 0.199222346114873, 0.941324937580778,
1, 0.971438368782348, 1, 1, 0.261050876294526, NaN, 0.00113648663275246,
NaN, NaN, 0.953445065176909, 5.49161967291152e-05, NaN, 0.000590014543772203,
0.999982700299285, 0.00135296027708626, 1, 0.0409008184478508,
NaN, 0.891996142719383, 0.00572915878256989, 0.995872043743129,
1, NaN, NaN, NaN, 0.993813065639553, NaN, NaN, NaN, NaN, 0.333333333333333,
NaN, 0.462998223209947, 0.0465549348230912, 0.797194659694581,
NaN, 0.433386754658534, 0.333333333333333, 0.0340277269556374,
NaN, 1, NaN, 0.945830148625487, 0.975005294668752, 1, NaN, 0.827886840316451,
0.90024391773039, 0.794054544390783, NaN, 0.436583079997355,
0.912449278688986, 0.152910279633107, 1.72997007151776e-05, 0.852983770022535,
NaN, NaN, 0.144497135806673, NaN, NaN, 0.05303517597611, NaN,
0.245671533948344, 0.204684590999682, 1, 0.00196488163927268,
NaN, 1, 0.994350282485876, NaN, 0.00156505026194397, NaN, 1,
0.1767290479113, 1, 0.5, NaN, NaN, NaN, NaN, NaN, NaN)), row.names = c(NA,
-100L), class = c("data.table", "data.frame"))
In case this makes things easier here are the packages I am using for this:
library(ggplot2)
library(tidyverse)
library(scales)
library("RColorBrewer")
library(ggsci)
In my original data frame (full data - too long to post here) there are many groups so it gets hard to distinguish the colors between the groups. These plots are just for data exploration so I would like to add prey name labels on top on my bar plots to be able to see the groups easier.
This is my original plot using ggplot:
ggplot(example, aes(x = factor(MONTH), y = BMASSprop, fill = prey_name))
geom_bar(stat="identity", position="fill")
ggtitle("Proportion of Prey Biomass in Adult YP Stomachs")
labs(x="Month", fill = "Prey Name")
labs(y = "Prey Biomass Consumed",
caption = "Source: DNR Diet Data")
scale_x_discrete(labels=c("June","July","August","November"))
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.ticks.length = unit(0.2,"cm"))
scale_fill_igv(palette = "default")
Again, in this plot the different groups (prey_names) are not that challenging to distinguish but with my additional groups some of the colors get too similar and hard to tell apart. Is there a way I can add labels (the prey names) on top of each unique color on the bar plot?
I have tried the following:
ggplot(example, aes(x = factor(MONTH), y = BMASSprop, fill = prey_name))
geom_bar(stat="identity", position="fill")
ggtitle("Proportion of Prey Biomass in Adult YP Stomachs")
labs(x="Month", fill = "Prey Name")
labs(y = "Prey Biomass Consumed",
caption = "Source: DNR Diet Data")
scale_x_discrete(labels=c("June","July","August","November"))
scale_y_continuous(labels = prey_name)
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.ticks.length = unit(0.2,"cm"))
scale_fill_igv(palette = "default")
This gives the following error:
#Error in check_breaks_labels(breaks, labels) : object 'prey_name' not found
I also tried this:
ggplot(example, aes(x = factor(MONTH), y = BMASSprop, fill = prey_name))
geom_bar(stat="identity", position="fill")
ggtitle("Proportion of Prey Biomass in Adult YP Stomachs")
labs(x="Month", fill = "Prey Name")
labs(y = "Prey Biomass Consumed",
caption = "Source: DNR Diet Data")
scale_x_discrete(labels=c("June","July","August","November"))
geom_text(aes(label = prey_name))
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.ticks.length = unit(0.2,"cm"))
scale_fill_igv(palette = "default")
This one gets me closer to what I want but there are duplicate labels and some are also stacked on top of each other.
So then I tried this:
ggplot(example, aes(x = factor(MONTH), y = BMASSprop, fill = prey_name))
geom_bar(stat="identity", position="fill")
ggtitle("Proportion of Prey Biomass in Adult YP Stomachs")
labs(x="Month", fill = "Prey Name")
labs(y = "Prey Biomass Consumed",
caption = "Source: DNR Diet Data")
scale_x_discrete(labels=c("June","July","August","November"))
geom_text(aes(label = prey_name), position = "stack")
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.ticks.length = unit(0.2,"cm"))
scale_fill_igv(palette = "default")
This completely messes up my plot.
Then I tried this:
ggplot(example, aes(x = factor(MONTH), y = BMASSprop, fill = prey_name))
geom_bar(stat="identity", position="fill")
ggtitle("Proportion of Prey Biomass in Adult YP Stomachs")
labs(x="Month", fill = "Prey Name")
labs(y = "Prey Biomass Consumed",
caption = "Source: DNR Diet Data")
scale_x_discrete(labels=c("June","July","August","November"))
geom_text(aes(label = prey_name), position = position_dodge(0.9))
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.ticks.length = unit(0.2,"cm"))
scale_fill_igv(palette = "default")
This still results in duplicate and stacked labeling.
Is there a way to add a single label for each prey name by month?
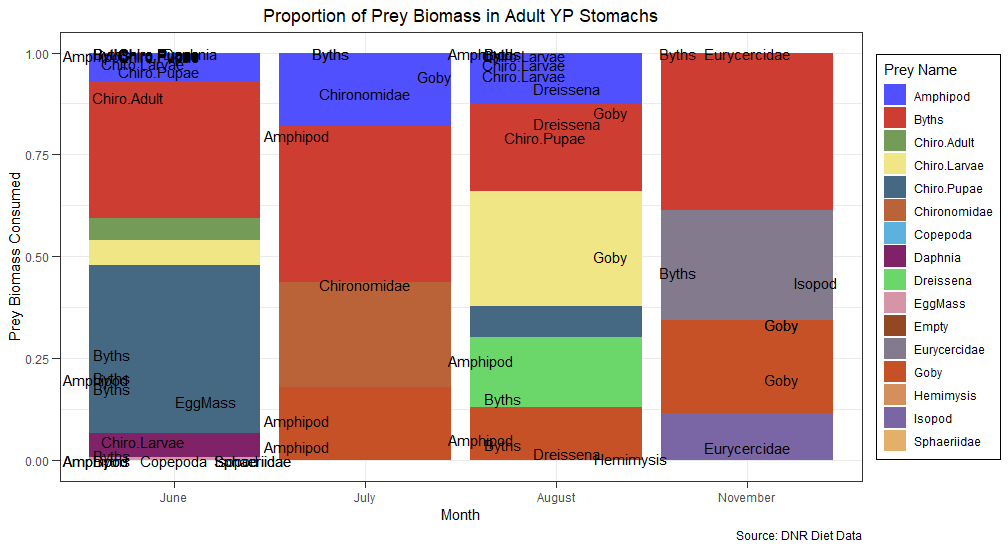
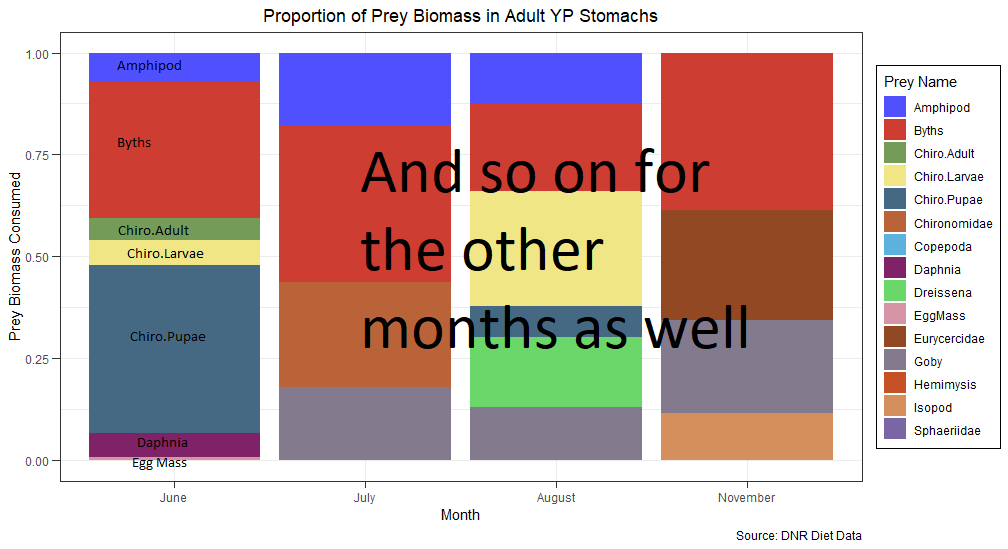
I would like to achieve something like this if possible:
Any suggestions on how to achieve this? Thank you for your time!
CodePudding user response:
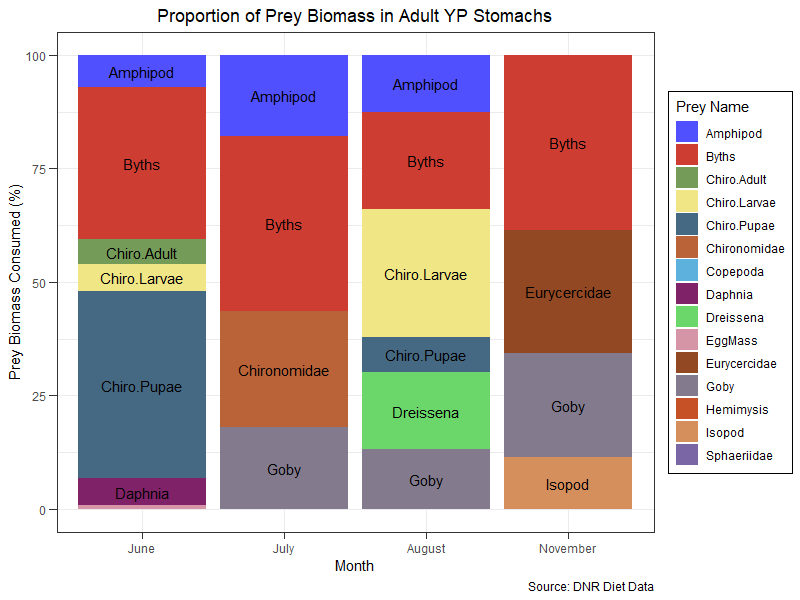
A way to achieve this, is to create a intermediate plot in order to summarize your data per prey to clean the data of the multiple names and frequencies:
clean_df = example %>% group_by(MONTH, prey_name) %>%
na.omit %>% summarize(prey_tot = sum(BMASSprop)) %>%
mutate(month_tot = sum(prey_tot, na.rm = T),
prey_frq = prey_tot * 100 / month_tot,
pos_label = sum(prey_frq, na.rm = T)-(cumsum(prey_frq)-0.5*prey_frq))
In the summarize you collapse all the same prey, then create the frequency in % of each prey per month and finally add the "pos_label" column which computes the y value at which you want your label.
Then you can generate the plot:
ggplot(clean_df, aes(as.factor(MONTH), prey_frq, fill=prey_name))
geom_bar(stat = "identity")
geom_text(data = subset(clean_df, prey_frq>5), aes(y = pos_label, label = prey_name))
ggtitle("Proportion of Prey Biomass in Adult YP Stomachs")
labs(x="Month", fill = "Prey Name", y = "Prey Biomass Consumed (%)", caption = "Source: DNR Diet Data")
scale_x_discrete(labels=c("June","July","August","November"))
scale_fill_igv(palette = "default")
theme_bw()
theme(legend.position = "right", plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.ticks.length = unit(0.2,"cm"))
You can set the value to filter out the label you want to see on the graph (here all prey with > 5% of total biomass)