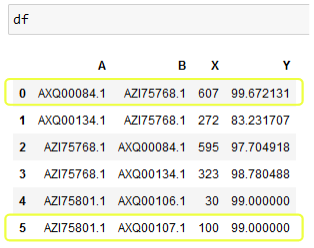
df = pd.DataFrame({'A':['AXQ00084.1', 'AXQ00134.1', 'AZI75768.1', 'AZI75768.1','AZI75801.1','AZI75801.1'],
'B':['AZI75768.1', 'AZI75768.1', 'AXQ00084.1', 'AXQ00134.1','AXQ00106.1','AXQ00107.1'],
'X': [607, 272, 595, 323,30,100],
'Y':[99.67213115, 83.23170732, 97.70491803, 98.7804878,99,99]})
I want to return the pairs of A and B which have the largest values of X and Y. For the df provided it would be the first and the last row. In the case where a column's value repeats e.g. A,B: AZI75801.1, AXQ00106.1 and AXQ00106.1, AXQ00107.1, return the pair AZI75801.1, AXQ00107.1 as they have the largest X and Y value.
How I'm attempting it:
- Find combinations of A and B which have already appeared but are swapped
df2 = df.assign(swap=df.apply(lambda r: ((df.loc[(df.A.eq(r.B)&df.B.eq(r.A))].index.values)<r.name).any(), axis=1))
- Split into 2 tables based on pairs of values that have been seen before vs. not
df_1=df2[df2.swap]
df_0=df2[~df2.swap]
df_1=df_1.sort_values(['A','X','Y'],ascending=False).groupby('A').first()
df_0=df_0.sort_values(['A','X','Y'],ascending=False).groupby('B').first()
df_1.reset_index(inplace=True)
df_0.reset_index(inplace=True)
df_1_grp=df_1.sort_values(['A','X','Y'],ascending=False).groupby('A').first().reset_index()
df_0_grp=df_0.sort_values(['A','X','Y'],ascending=False).groupby('A').first().reset_index()
merge_df=pd.merge(df_1_grp, df_0_grp, left_on= ['A', 'B'],
right_on= ['B', 'A'],
how='outer',suffixes=('_1','_2'))
- Grouping to get the max value and then merging which gives:
A_1 B_1 X_1 Y_1 swap_1 A_2 B_2 X_2 Y_2 swap_2
0 AZI75768.1 AXQ00084.1 595.0 97.704918 True NaN NaN NaN NaN NaN
1 NaN NaN NaN NaN NaN AXQ00134.1 AZI75768.1 272.0 83.231707 False
2 NaN NaN NaN NaN NaN AZI75801.1 AXQ00107.1 100.0 99.000000 False
But this still doesn't solve my problem as it returns the pair AXQ00134.1, AZI75768.1 which is incorrect as AZI75768.1 has the highest X,Y value with AZI75768.1.
Any help is greatly appreciated.
CodePudding user response:
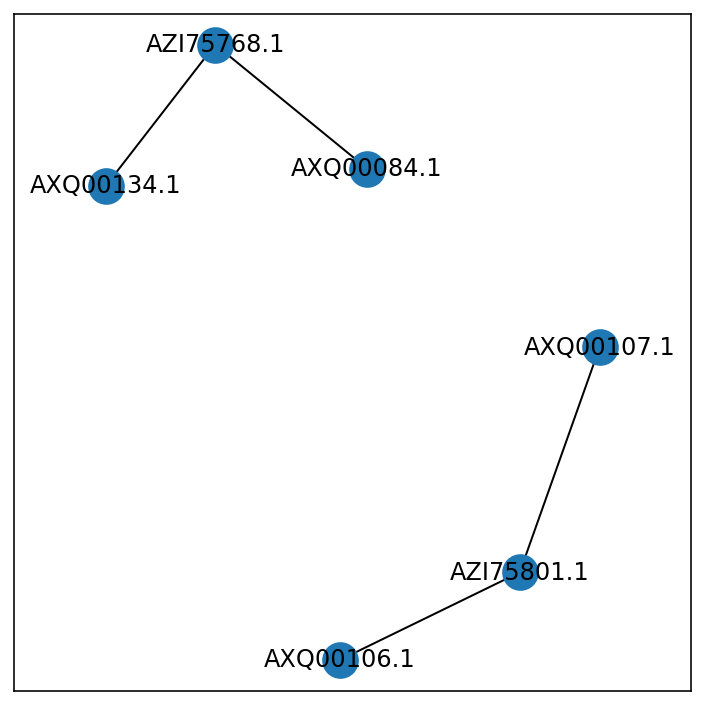
IIUC, this is a graph problem. You have connected components and want to find the max of the disjoint subgraphs.
You can use networkx to identify the disjoint subgraphs and use this to form groups for pandas' groupby:
import networkx as nx
G = nx.from_pandas_edgelist(df, source='A', target='B')
group = {k: v for v,s in enumerate(nx.connected_components(G)) for k in s}
# {'AZI75768.1': 0, 'AXQ00084.1': 0, 'AXQ00134.1': 0,
# 'AXQ00106.1': 1, 'AXQ00107.1': 1, 'AZI75801.1': 1}
out = (df
.sort_values(by=['X', 'Y'])
.groupby(df['A'].map(group))
.last()
)
Output:
A B X Y
A
0 AXQ00084.1 AZI75768.1 607 99.672131
1 AZI75801.1 AXQ00107.1 100 99.000000
Your graph:
CodePudding user response:
double_max = []
for (a, b), grouped_data in df.groupby(['A', 'B'])
x_max_index= np.argmax(grouped_data['X'])
y_max_index= np.argmax(grouped_data['Y'])
if x_max_index == y_max_index:
max_row = grouped_data.iloc[x_max_index]
double_max.append([a, b, max_row ['X'], max_row['Y'])