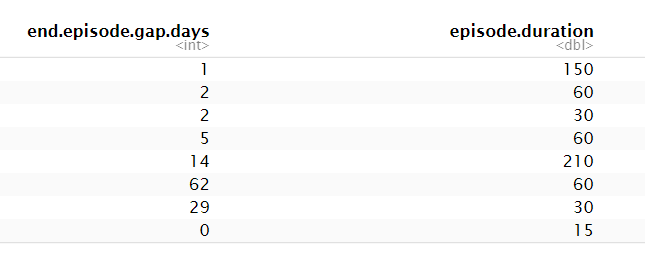
I am currently working on a data set in which I am trying to track patients using a medication. I have the data in episodes like this
So basically this data says that in the first episode the patient used the medication for 150 days then the patient stopped for 1 day then the patient used the medication for 60 days then stopped for 2 days etc.
What I want to do is to convert this to longitudinal data so I have the: 150 1s then a single 0 then sixty 1s then 2 zeros etc.
How to do that?
CodePudding user response:
data<-data.frame(
end.episode.gap.days= c(1,2,2, .....),
episode.duration = c(150,60,30,.....))
values <- unlist(lapply(1:NROW(data), function(i){
c(rep(1,data$episode.duration[i]),rep(0,data$end.episode.gap.days[i]))} ))
print(values)
You must complete the numbers in .....
CodePudding user response:
This is a slightly modified version of Ricardo's answer that should be able to "carry over" any id columns:
data <- data.frame(
id = 1:8,
name = letters[1:8],
end.episode.gap.days = c(
1, 2, 2, 5, 14, 62, 29, 0
),
episode.duration = c(
150,
60,
30,
60,
210,
60,
30,
15
)
)
result <- do.call(rbind, lapply(1:nrow(data), function(i) {
longitudinal_values <- c(rep(1, data$episode.duration[i]), rep(0, data$end.episode.gap.days[i]))
id_cols <- data[rep(i, length(longitudinal_values)), !(names(data) %in% c("end.episode.gap.days", "episode.duration"))]
cbind(id_cols, longitudinal_values)
}))