i am currently plotting (long format) data which consists of fluorescence (RFU) on the 1. Y-Axis and Growth (OD600) on the 2. Y-Axis. I have managed to create the plots, but i find it very difficult to log transform the 2. Y-axis (for OD600) and not messing up the entire plot. (The data is all derived from the same data frame)
My question is this: Is there any way to log10 transform only the 2. Y-axis (from 0.01-1) and making perhaps 5 breaks something like:("0.01","0.1","0.5","0.1")?
My code looks like this: (i apologize for ugly code)
for (i in 1:length(unique(lf_combined$media)[grepl("^. (gfp)$",unique(lf_combined$media))])){
print(i)
coeff <- 1/max(lf_combined_test$normalized_gfp)
p1<-lf_combined_test[lf_combined_test$media %in% unique(lf_combined$media)[grepl("^. (gfp)$",unique(lf_combined$media))][i], ] %>%
# filter(normalized_gfp>0) %>%
filter(row_number() %% 3 == 1) %>%
ggplot( aes(x=time))
geom_bar( aes(y=normalized_gfp), stat="identity", size=.1, fill="green", color="green", alpha=.4)
geom_line( aes(y=od / coeff), size=2, color="tomato")
scale_x_continuous(breaks = round(seq(0,92, by = 5),1))
geom_vline(xintercept = 12, linetype="dotted",
color = "blue", size=1)
scale_y_continuous(limits = c(0,80000),
name = "Relative Flourescence [RFU]/[OD] ",
sec.axis = sec_axis(~.*coeff, name="[OD600]")
)
scale_y_log10(limits=c(0.01,1))
theme_grey()
theme(
axis.title.y = element_text(color = "green", size=13),
axis.title.y.right = element_text(color = "tomato", size=13)
)
ggtitle(paste("Relative fluorescence & OD600 time series for",unique(lf_combined$media)[grepl("^. (gfp)$",unique(lf_combined$media))][i],sep=" "))
print(p1)
)
}
Which gives a plots that looks like this for now:
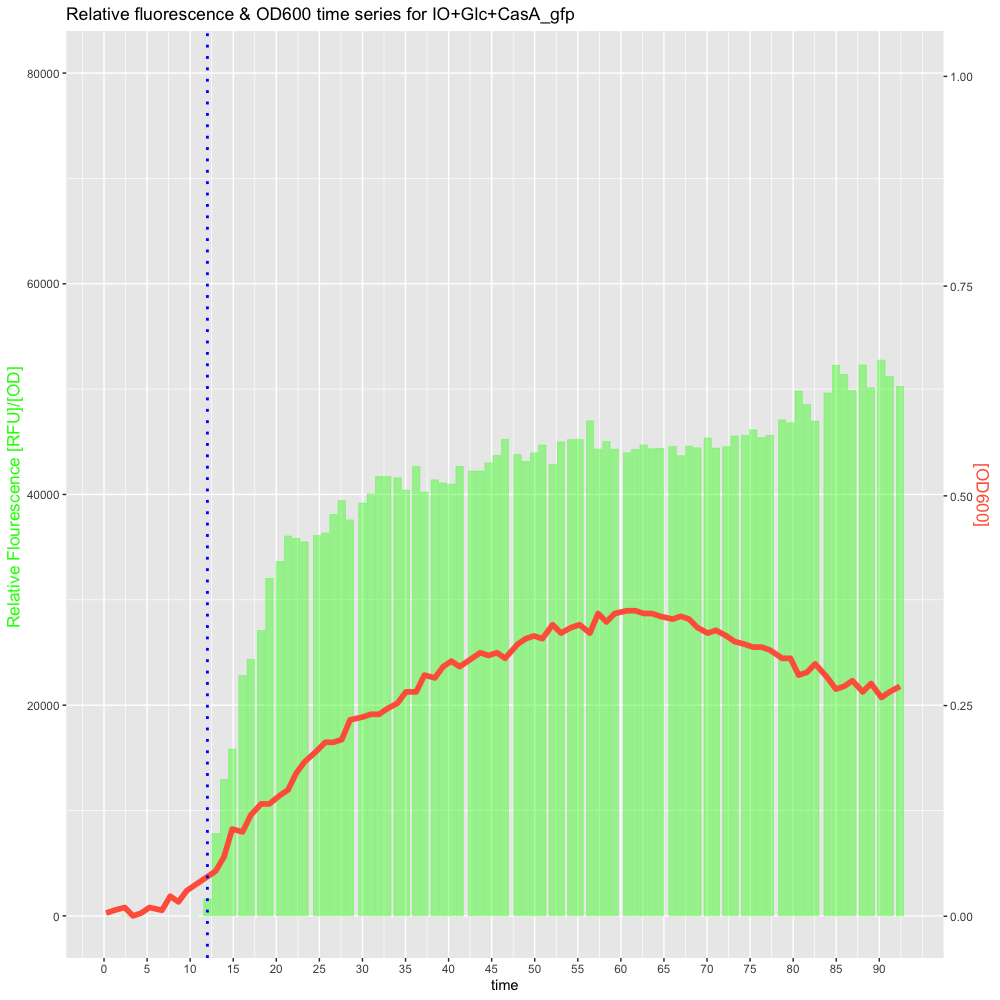
Thank you very much in advance! :))
CodePudding user response:
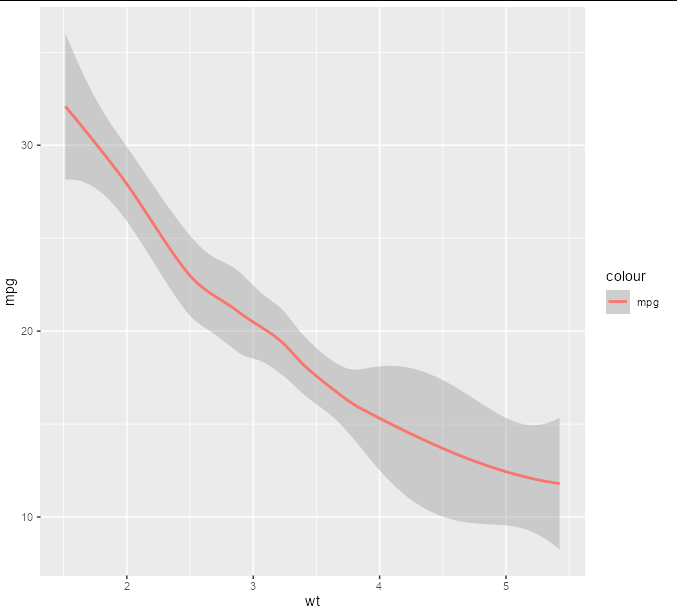
Yes, this is certainly possible. Without your data set it is difficult to give you specific code, but here is an example using the built-in mtcars data set. We plot a best-fitting line for mpg against an x axis of wt.
p <- ggplot(mtcars, aes(wt, mpg)) geom_smooth(aes(color = 'mpg'))
p
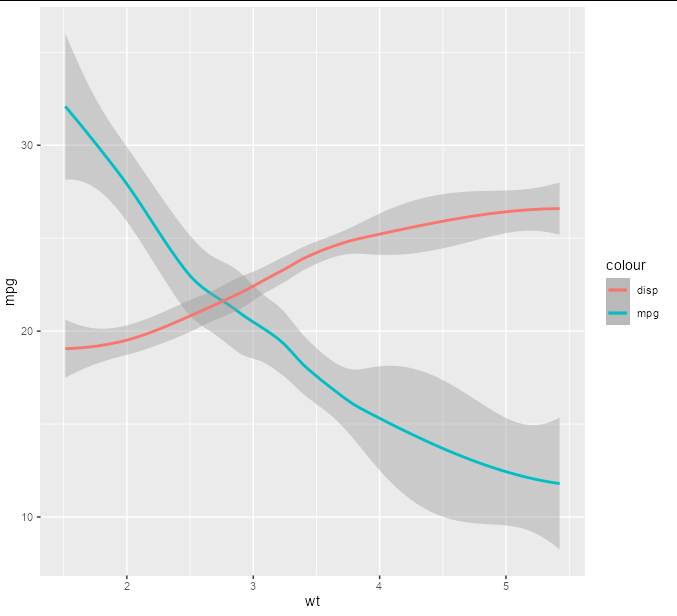
Suppose we want to draw the value of disp according to a log scale which we will show on the y axis. We need to carry out the log transform of our data to do this, but also multiply it by 10 to get it on a similar visual scale to the mpg line:
p <- p geom_smooth(aes(y = 10 * log10(disp), color = 'disp'))
p
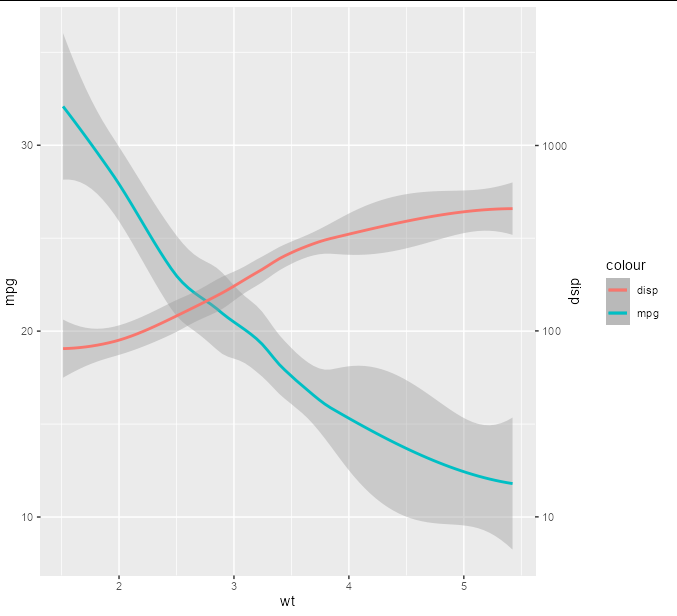
To draw the secondary axis in, we need to supply it with the reverse transformation of 10 * log10(x), which is 10^(x/10), and we will supply appropriately logarithmic breaks at 10, 100 and 1000
p scale_y_continuous(
sec.axis = sec_axis(~ 10^(.x/10), breaks = c(10, 100, 1000), name = 'disp'))
It seems that you are generating the values of your line by using od / coeff, and reversing that transform with .*coeff, which seems appropriate, but to get a log10 axis, you will need to do something like log10(od) * constant and reverse it with 10^(od/constant). Without your data, it's impossible to know what this constant should be, but you can play around with different values until it looks right visually.