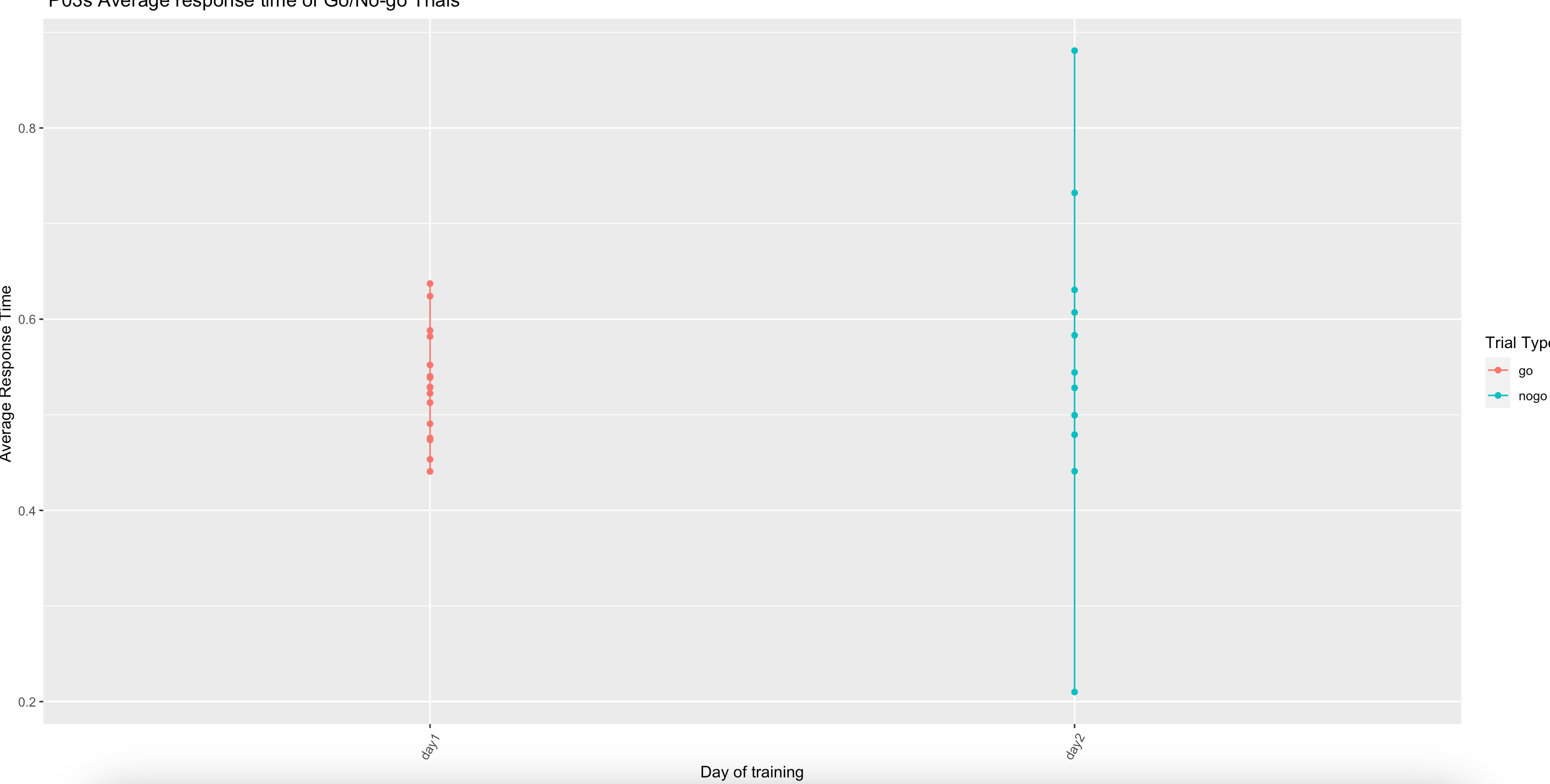
I am trying to create a line graph which shows the response times of a go no go trial over a few weeks. However, instead of having the dates on the x axis, I just wanted it to display in the order like 'day 1, day 2' etc... After following advice from another user I added this to my ggplot code
mutate(days = paste0("day",row_number()))
The full ggplot code is as follows:
p03_dropped_rt %>%
mutate(days = paste0("day",row_number())) %>%
ggplot(aes(x = fct_inorder(days), y = mean, group = trialtype))
geom_line(aes(color = trialtype))
geom_point(aes(color = trialtype))
theme(axis.text.x = element_text(angle = 60, vjust = 0.5))
labs(title=" P03s Average response time of Go/No-go Trials",
x = "Day of training",
y = "Average Response Time ",
color = "Trial Type")
I thought this would change the dates to days and put them in order, however, the produced visualisation is wrong (See picture for clarification)
I am just wondering if anyone has any ideas on how to fix this?
here is the data:
structure(list(day = structure(c(18880, 18880, 18908, 18908,
18911, 18912, 18912, 18913, 18913, 18914, 18914, 18915, 18915,
18916, 18916, 18917, 18917, 18918, 18918, 18919, 18920, 18921,
18921, 18922, 18922, 18923, 18924), class = "Date"), trialtype = c("go",
"nogo", "go", "nogo", "go", "go", "nogo", "go", "nogo", "go",
"nogo", "go", "nogo", "go", "nogo", "go", "nogo", "go", "nogo",
"go", "go", "go", "nogo", "go", "nogo", "go", "go"), mean = c(0.453363414634146,
0.21005, 0.63720350877193, 0.8809, 0.624077192982456, 0.581956842105263,
0.63065, 0.588196842105263, 0.607075, 0.540253289473684, 0.73215,
0.529153097345133, 0.583133333333333, 0.538794736842105, 0.4409,
0.522421428571429, 0.4792, 0.552129824561403, 0.5282, 0.512742105263158,
0.473664912280702, 0.528580701754386, 0.544266666666667, 0.490643859649123,
0.49955, 0.440738596491228, 0.475708771929825)), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -27L), groups = structure(list(
day = structure(c(18880, 18908, 18911, 18912, 18913, 18914,
18915, 18916, 18917, 18918, 18919, 18920, 18921, 18922, 18923,
18924), class = "Date"), .rows = structure(list(1:2, 3:4,
5L, 6:7, 8:9, 10:11, 12:13, 14:15, 16:17, 18:19, 20L,
21L, 22:23, 24:25, 26L, 27L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -16L), .drop = TRUE))
CodePudding user response:
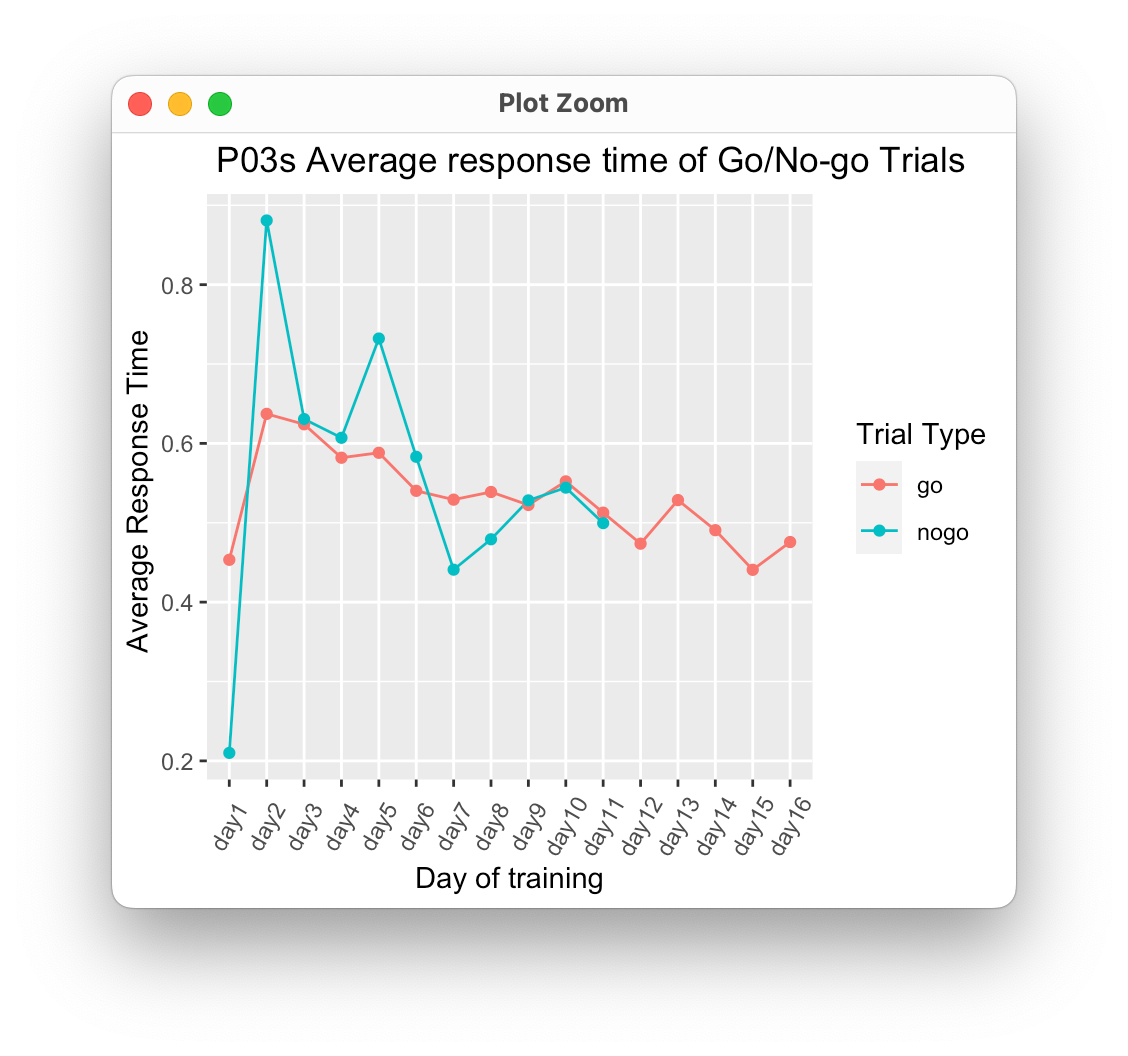
It looks like the p03_dropped_rt was grouped by day earlier, so when you applied your mutate, it labeled each "go" trial as day1 and each "nogo" trial as day2. Instead, we can group by trialtype, arrange by day, and then mutate:
p03_dropped_rt %>%
group_by(trialtype) %>%
arrange(day) %>%
mutate(days = paste0("day",row_number())) %>%
ggplot(aes(x = fct_inorder(days), y = mean, group = trialtype))
geom_line(aes(color = trialtype))
geom_point(aes(color = trialtype))
theme(axis.text.x = element_text(angle = 60, vjust = 0.5))
labs(title=" P03s Average response time of Go/No-go Trials",
x = "Day of training",
y = "Average Response Time ",
color = "Trial Type")