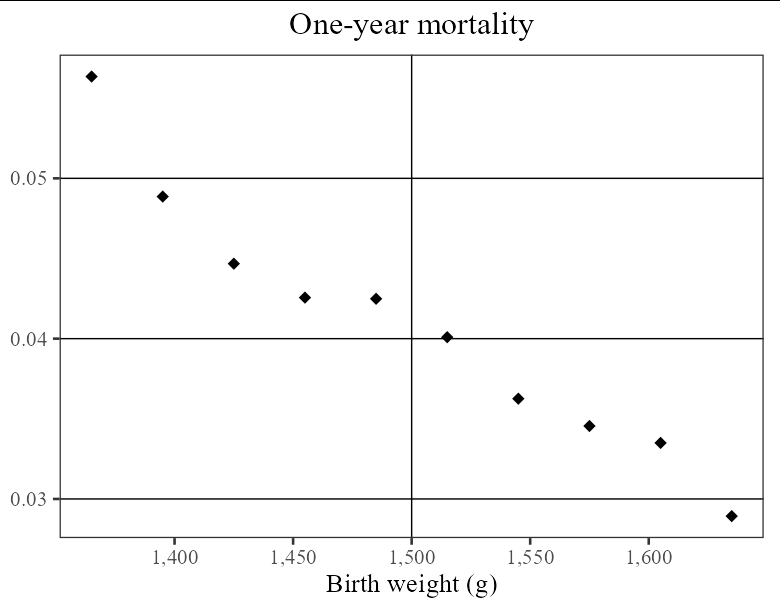
so basically I have created the bins and the have the means of each bin, having these two columns in a dataframe. Now I am plotting these two columns, but I want the exact number as x lable instead of bins. I am considering renaming each bin by its mid-point. please look at the pictures. The first one is my current plot and the second is the plot I want to acheive.
my current plot:
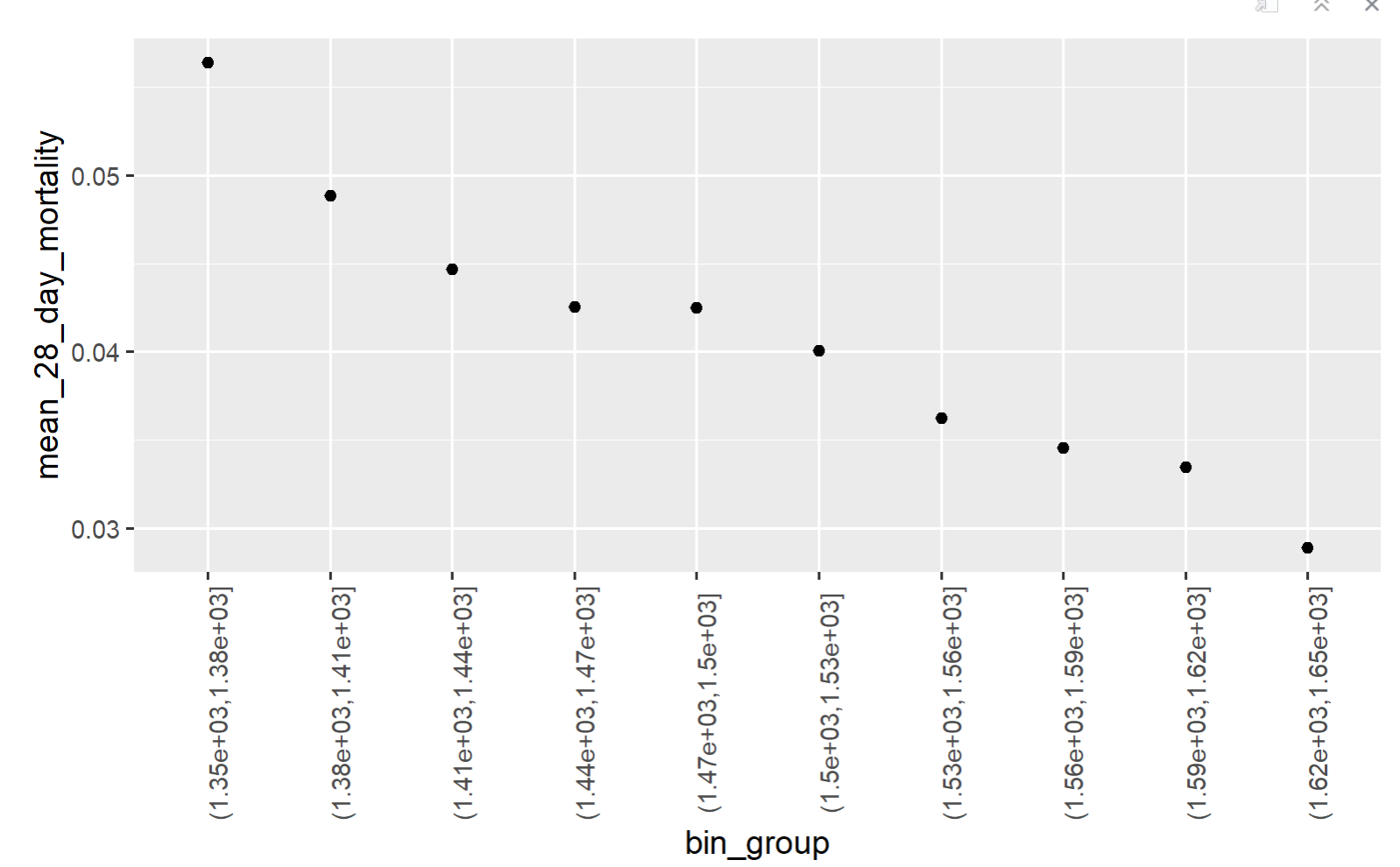 what I want to have:
what I want to have:
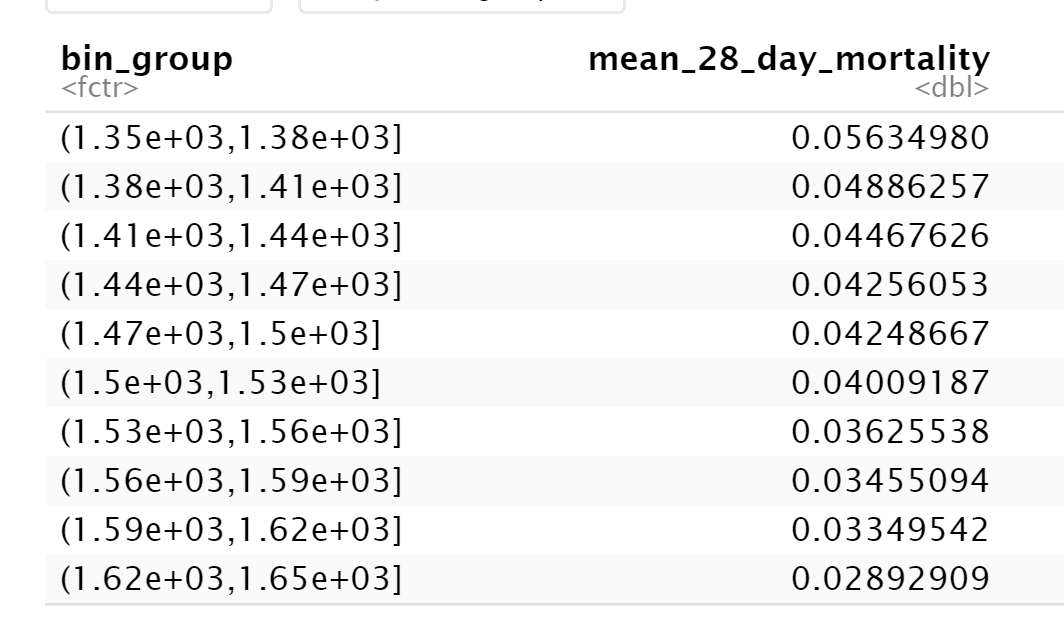
 my data frame is like this:
my data frame is like this:
CodePudding user response:
If you have groups that (I assume) you made with cut, you could pull out the max and min and then calc the mean before you summarize and plot. Note that I made the regex pretty long because I don't personally know if cut always makes left or inclusive or exclusive.
library(tidyverse)
#example like yours
mtcars |>
mutate(grp = cut(hp, 10)) |>
group_by(grp) |>
summarise(mpg_mean = mean(mpg)) |>
ggplot(aes(grp, mpg_mean))
geom_point()
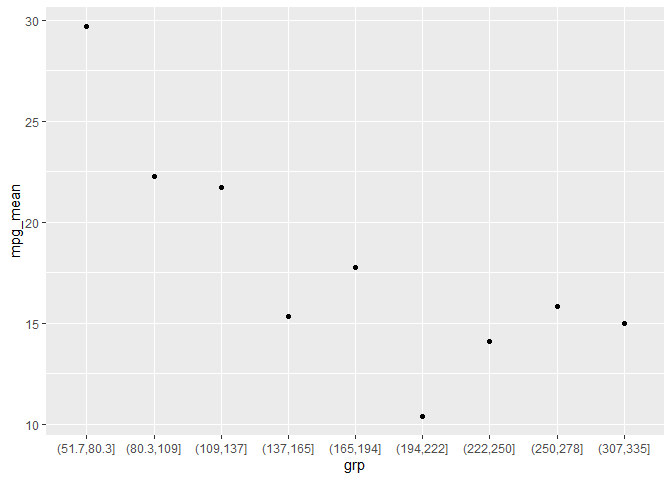
#solution
mtcars |>
mutate(grp = cut(hp, 10)) |>
extract(grp,
into = c("min", "max"),
remove = FALSE,
regex = "(?:\\(|\\[)(.*),(.*)(?:\\)|\\])",
convert = TRUE) |>
mutate(mean_grp = (min max)/2)|>
group_by(mean_grp) |>
summarise(mpg_mean = mean(mpg)) |>
ggplot(aes(mean_grp, mpg_mean))
geom_point()
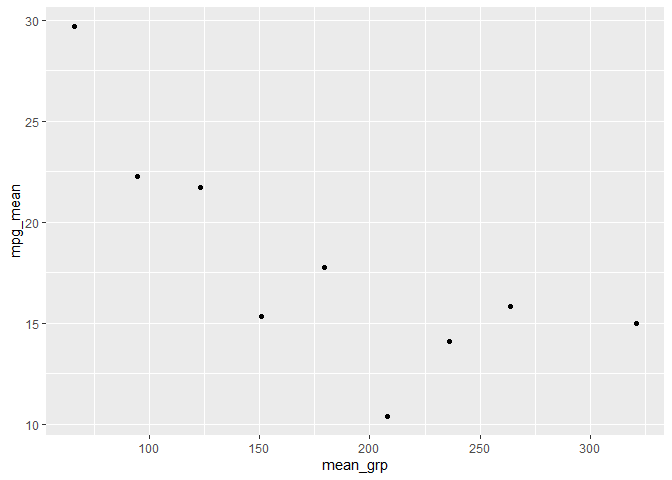
CodePudding user response:
To reproduce the style of the plot image you included, you can do:
library(tidyverse)
df %>%
mutate(bin_group = gsub("\\(|\\]", "", bin_group)) %>%
separate(bin_group, sep = ",", into = c("lower", "upper")) %>%
mutate(across(lower:upper, as.numeric)) %>%
mutate(`Birth weight (g)` = (upper lower) / 2) %>%
ggplot(aes(`Birth weight (g)`, mean_28_day_mortality))
geom_vline(xintercept = 1500)
geom_point(shape = 18, size = 4)
scale_x_continuous(labels = scales::comma)
labs(title = "One-year mortality", y = NULL)
theme_bw(base_family = "serif", base_size = 20)
theme(panel.grid.major.x = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major.y = element_line(color = "black", size = 0.5),
plot.title = element_text(hjust = 0.5))
Data used (obtained from image in question using OCR)
df <- structure(list(bin_group = structure(1:10,
levels = c("(1.35e 03,1.38e 03]",
"(1.38e 03,1.41e 03]", "(1.41e 03,1.44e 03]", "(1.44e 03,1.47e 03]",
"(1.47e 03,1.5e 03]", "(1.5e 03,1.53e 03]", "(1.53e 03,1.56e 03]",
"(1.56e 03,1.59e 03]", "(1.59e 03,1.62e 03]", "(1.62e 03,1.65e 03]"
), class = "factor"), mean_28_day_mortality = c(0.0563498, 0.04886257,
0.04467626, 0.04256053, 0.04248667, 0.04009187, 0.03625538, 0.03455094,
0.03349542, 0.02892909)), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -10L))