First we prepare some toy data that sufficiently resembles the one I am working with.
rawdata <- data.frame(Score = rnorm(1000, seq(1, 0, length.out = 10), sd = 1),
Group = rep(LETTERS[1:3], 10000))
stdev <- c(10.78,10.51,9.42)
Now we plot the estimated densities via geom_density_ridges. I also add a grey highlight around zero via geom_rect. I also flip the chart with coord_flip.
p <- ggplot(rawdata, aes(x = Score, y = Group))
scale_y_discrete()
geom_rect(inherit.aes = FALSE, mapping = aes(ymin = 0, ymax = Inf, xmin = -0.1 * min(stdev), xmax = 0.1 * max(stdev)),
fill = "grey", alpha = 0.5)
geom_density_ridges(aes(fill = Group), scale = 0.5, size = 1, alpha=0.5)
scale_color_manual(values = col)
scale_fill_manual(values = col)
labs(title="Toy Graph", y="Group", x="Value")
coord_flip(xlim = c(-8, 8), ylim = NULL, expand = TRUE, clip = "on")
p
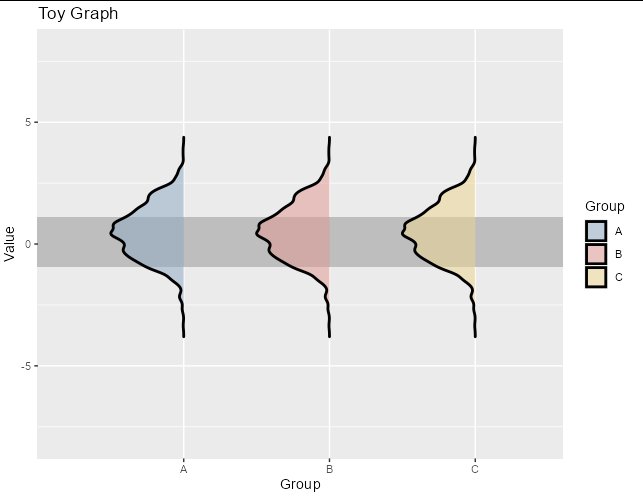
 And this is the solution I get, which is close to what I was expecting, despite the detail of this enormous gap between the y axis an the start of the first factor in the x axis
And this is the solution I get, which is close to what I was expecting, despite the detail of this enormous gap between the y axis an the start of the first factor in the x axis A. I tried using expand=c(0,0) inside scale_y_discrete() following some suggestions from other posts, but it does not make the gap smaller at all. If possible I would still like to have a certain gap, although minimal. I've been also trying to flip the densities in the y axis so the gap is filled by first factor density plot but I have been unsuccessful as it does not seem as trivial as one could expect.
Sorry, I know this might be technically two different questions, "How to reduce the gap from the y axis to the first density plot?" and "How to flip the densities from y axis to reduce the gap?" But I would really be happy with the first one as I understand the second question seems to be apparently less straightforward.
Thanks in advance! Any help is appreciated.
CodePudding user response:
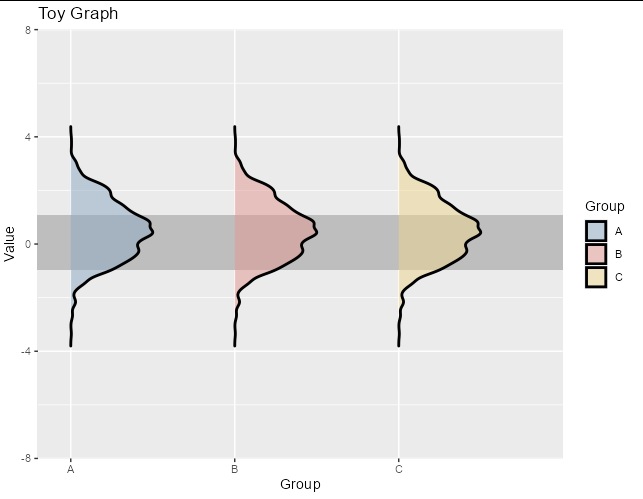
Flipping the densities also effectively reduces the space, so this might be all you need to do. You can achieve it with a negative scale parameter:
ggplot(rawdata, aes(x = Score, y = Group))
scale_y_discrete()
geom_rect(inherit.aes = FALSE,
mapping = aes(ymin = 0, ymax = Inf,
xmin = -0.1 * min(stdev),
xmax = 0.1 * max(stdev)),
fill = "grey", alpha = 0.5)
geom_density_ridges(aes(fill = Group), scale = -0.5, size = 1, alpha = 0.5)
scale_color_manual(values = col)
scale_fill_manual(values = col)
labs(title = "Toy Graph", y = "Group", x = "Value")
coord_flip(xlim = c(-8, 8), ylim = NULL, expand = TRUE, clip = "on")
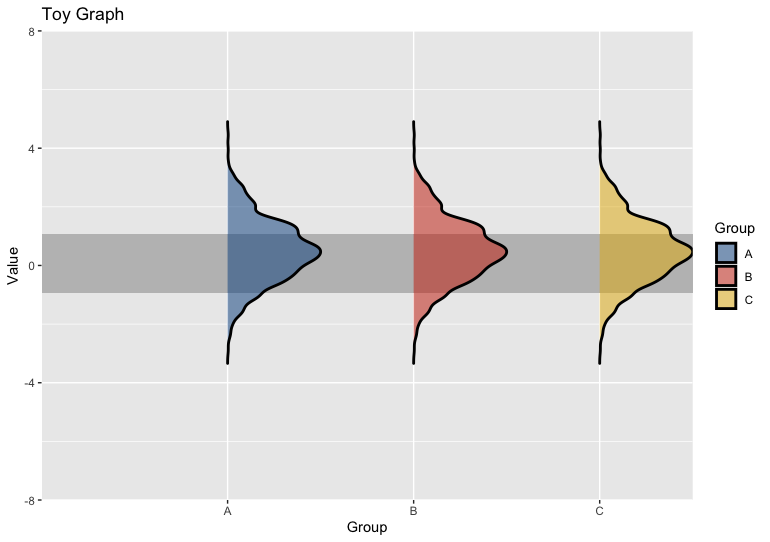
If you want to keep the densities pointing the same way but just reduce space on the left side, simply set hard limits in your coord_flip, with no expansion:
ggplot(rawdata, aes(x = Score, y = Group))
geom_rect(inherit.aes = FALSE,
mapping = aes(ymin = 0, ymax = Inf,
xmin = -0.1 * min(stdev),
xmax = 0.1 * max(stdev)),
fill = "grey", alpha = 0.5)
geom_density_ridges(aes(fill = Group), scale = 0.5, size = 1, alpha = 0.5)
scale_color_manual(values = col)
scale_fill_manual(values = col)
scale_y_discrete()
labs(title = "Toy Graph", y = "Group", x = "Value")
coord_flip(xlim = c(-8, 8), ylim = c(0.8, 4), expand = FALSE)