I have this line of code:
df = pd.read_csv('some_file.txt',engine ='python',
delimiter = '\t', header=None, encoding="utf-16")
I'm using those txt files quiet often in my lab, one of our machines gives them as output. If I only use the delimiter I get a nice table, but with the first element as header for everything. If I only use header = None I get rid of the header, but have a bunch of \t everywhere. If I try to use both commands, I get this error:
ParserError: Expected 1 fields in line 3, saw 23
When removing enigne = 'python' I get a similar error. (also tried seperator and a bunch of other things)
Help would be very much appreciated!
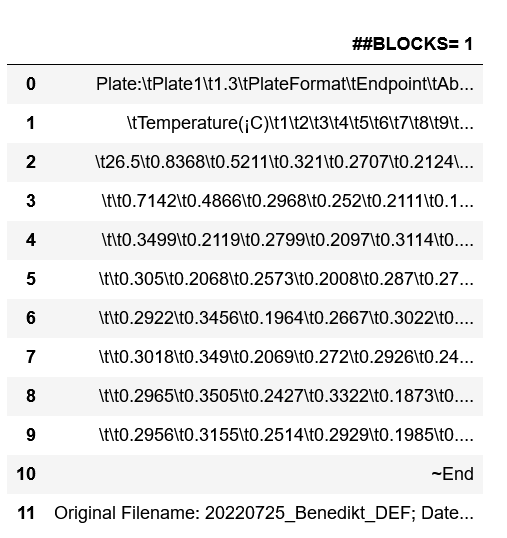
Edit: As requested that's how the file looks like:
##BLOCKS= 1
Plate: Plate1 1.3 PlateFormat Endpoint Absorbance Raw FALSE 1 1 562 1 12 96 1 8
Temperature(¡C) 1 2 3 4 5 6 7 8 9 10 11 12
26.5 0.8368 0.5211 0.321 0.2707 0.2124 0.1768 0.1694 0.1635 0.1659 0.1029 0.1032 0.104
0.7142 0.4866 0.2968 0.252 0.2111 0.1737 0.1633 0.162 0.1599 0.1009 0.1007 0.1025
0.3499 0.2119 0.2799 0.2097 0.3114 0.3393 0.2544 0.2965 0.2392 0.3063 0.3093 0.2655
0.305 0.2068 0.2573 0.2008 0.287 0.2765 0.2373 0.2703 0.2357 0.2865 0.2926 0.263
0.2922 0.3456 0.1964 0.2667 0.3022 0.2596 0.2256 0.2387 0.2498 0.2936 0.2396 0.3411
0.3018 0.349 0.2069 0.272 0.2926 0.2444 0.2141 0.2348 0.2486 0.2678 0.2346 0.2944
0.2965 0.3505 0.2427 0.3322 0.1873 0.2286 0.3758 0.208 0.3023 0.3573 0.3141 0.2658
0.2956 0.3155 0.2514 0.2929 0.1985 0.2379 0.1898 0.2101 0.3211 0.3558 0.3121 0.2567
~End
Original Filename: 20220725_Benedikt_DEF; Date Last Saved: 7/25/2022 2:31:30 PM
That's how it looks like when I read it without pandas:
['##BLOCKS= 1\n', 'Plate:\tPlate1\t1.3\tPlateFormat\tEndpoint\tAbsorbance\tRaw\tFALSE\t1\t\t\t\t\t\t1\t562 \t1\t12\t96\t1\t8\t\t\n', '\tTemperature(¡C)\t1\t2\t3\t4\t5\t6\t7\t8\t9\t10\t11\t12\t\t\n', '\t26.5\t0.8368\t0.5211\t0.321\t0.2707\t0.2124\t0.1768\t0.1694\t0.1635\t0.1659\t0.1029\t0.1032\t0.104\t\t\n', '\t\t0.7142\t0.4866\t0.2968\t0.252\t0.2111\t0.1737\t0.1633\t0.162\t0.1599\t0.1009\t0.1007\t0.1025\t\t\n', '\t\t0.3499\t0.2119\t0.2799\t0.2097\t0.3114\t0.3393\t0.2544\t0.2965\t0.2392\t0.3063\t0.3093\t0.2655\t\t\n', '\t\t0.305\t0.2068\t0.2573\t0.2008\t0.287\t0.2765\t0.2373\t0.2703\t0.2357\t0.2865\t0.2926\t0.263\t\t\n', '\t\t0.2922\t0.3456\t0.1964\t0.2667\t0.3022\t0.2596\t0.2256\t0.2387\t0.2498\t0.2936\t0.2396\t0.3411\t\t\n', '\t\t0.3018\t0.349\t0.2069\t0.272\t0.2926\t0.2444\t0.2141\t0.2348\t0.2486\t0.2678\t0.2346\t0.2944\t\t\n', '\t\t0.2965\t0.3505\t0.2427\t0.3322\t0.1873\t0.2286\t0.3758\t0.208\t0.3023\t0.3573\t0.3141\t0.2658\t\t\n', '\t\t0.2956\t0.3155\t0.2514\t0.2929\t0.1985\t0.2379\t0.1898\t0.2101\t0.3211\t0.3558\t0.3121\t0.2567\t\t\n', '\n', '~End\n', 'Original Filename: some_file; Date Last Saved: 7/25/2022 2:31:30 PM\n']
When I use just use the pd.read_csv(file, encoding =''utf-16')
I get this:
It' basically a file that is stating the wavelength absorbance from a sample plate with 8 rows and 12 columns (96 samples).
CodePudding user response:
Assuming all the files have the same structure and you only want the data; skip the first four rows, don't use the last three rows, whitespace delimiter, no header, python engine.
>>> df = pd.read_csv(csv,skiprows=4,skipfooter=3,header=None,delim_whitespace=True,engine='python')
>>> df
0 1 2 3 4 5 6 7 8 9 10 11
0 0.7142 0.4866 0.2968 0.2520 0.2111 0.1737 0.1633 0.1620 0.1599 0.1009 0.1007 0.1025
1 0.3499 0.2119 0.2799 0.2097 0.3114 0.3393 0.2544 0.2965 0.2392 0.3063 0.3093 0.2655
2 0.3050 0.2068 0.2573 0.2008 0.2870 0.2765 0.2373 0.2703 0.2357 0.2865 0.2926 0.2630
3 0.2922 0.3456 0.1964 0.2667 0.3022 0.2596 0.2256 0.2387 0.2498 0.2936 0.2396 0.3411
4 0.3018 0.3490 0.2069 0.2720 0.2926 0.2444 0.2141 0.2348 0.2486 0.2678 0.2346 0.2944
5 0.2965 0.3505 0.2427 0.3322 0.1873 0.2286 0.3758 0.2080 0.3023 0.3573 0.3141 0.2658
6 0.2956 0.3155 0.2514 0.2929 0.1985 0.2379 0.1898 0.2101 0.3211 0.3558 0.3121 0.2567