My data looks like this:
dput(NDVI)
structure(list(date = structure(c(19091, 19091, 19092, 19092,
19093, 19093, 19094, 19094, 19095, 19095, 19096, 19096, 19097,
19097, 19098, 19098, 19099, 19099, 19100, 19100, 19101, 19101,
19102, 19102, 19103, 19103, 19104, 19104, 19105, 19105, 19106,
19106, 19107, 19107, 19109, 19109, 19110, 19110, 19111, 19111,
19112, 19112, 19113, 19113, 19114, 19114), class = "Date"), target = c("grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree"), cNDVI = c(0.500272817021574,
0.696083797270416, 0.489691804085761, 0.692795403987576, 0.570071858811957,
0.698997926130435, 0.510441047568798, 0.693961708101642, 0.497010486993716,
0.677617144059372, 0.489045582825731, 0.672753166123319, 0.488193790481098,
0.663609030602538, 0.488885606064579, 0.644550512172325, 0.49094954276713,
0.625607907143415, 0.477036686933829, 0.606892335456545, 0.46818825238034,
0.598763281698822, 0.464516342335005, 0.594977691269838, 0.463896574547792,
0.611135472706338, 0.60754090771395, 0.657544930586677, 0.483871290250419,
0.641341913760879, 0.455301838422566, 0.625854396853959, 0.461233936815514,
0.620543555588439, 0.467959523995231, 0.615756464377253, 0.441352232545973,
0.597588129996248, 0.443084476959249, 0.588500193948734, 0.447515720605795,
0.569604003059877, 0.445609012507936, 0.562613166422831, 0.495220373276007,
0.58718847627638), TempAmb_Avg = c(13.16, NA, 7.929, NA, 12.29,
NA, 10.37, NA, 10.91, NA, 10.14, NA, 9.15, NA, 11.25, NA, 9.17,
NA, 11.94, NA, 11.26, NA, 9.45, NA, 9.09, NA, NA, NA, 6.447,
NA, 9.14, NA, 8.02, NA, 10.54, NA, 10.12, NA, 11.56, NA, 12.3,
NA, 10.82, NA, 11.17, NA), sd_NDVI = c(0.00206275442936463, 0.00483529298045758,
0.00143646028945461, 0.00547488157335041, 0.0107421213523848,
NA, 0.0057345829257413, 0.00239673774468361, 0.0122234699109748,
0.0041293261382514, 0.00159520722600412, 0.00400695247999584,
0.0014958900631327, 0.00207345410768067, 0.00142529228044798,
0.00188141954676642, 0.00254072590985734, 0.00374571468034617,
0.00162042982764391, 0.00371980695360771, 0.00149205232583476,
0.00860693732895818, 0.00195420993834865, 0.0135370828066329,
0.00267586232456702, 0.00366685362066545, 0.00548912106770043,
0.0115656879529442, 0.00892427653015911, 0.00876020639322138,
0.00793602431786174, 0.00308048721534673, 0.0022395875936781,
0.00464228152203431, 0.010403456112334, 0.01197424728207, 0.0203370856390955,
0.00842098123326728, 0.00179785152483323, 0.00238532744345886,
0.00174703441109131, 0.0016114072840488, 0.00150060553756978,
0.00245323735514388, 0.0276795273807586, 0.0187674033605546)), row.names = c(NA,
46L), class = "data.frame")
I'm trying to plot the standard deviation sd_NDVI on cNDVI in a two-way y-axis.
plot <- ggplot(NDVI)
geom_bar(aes(x=date, y=TempAmb_Avg),stat="identity",colour="blue", fill="blue")
geom_point(aes(x=date, y=30*cNDVI,color=target),stat="identity")
geom_line(aes(x=date, y=30*cNDVI,color=target),stat="identity")
geom_errorbar(aes(ymin=30*cNDVI-30*sd_NDVI, ymax=30*cNDVI 30*sd_NDVI, width=.2, position = position_dodge(0.05)),color=target, stat="identity")
labs(x="TIME",y="TºC") theme_bw()
plot
I'm getting this error:
Error in layer(data = data, mapping = mapping, stat = stat, geom = GeomErrorbar, :
object 'target' not found
and despite the plot being displayed the standard deviation sd_NDVI in cNDVI is not. How can I display the standard deviation in these conditions?
Thank you in advance!
CodePudding user response:
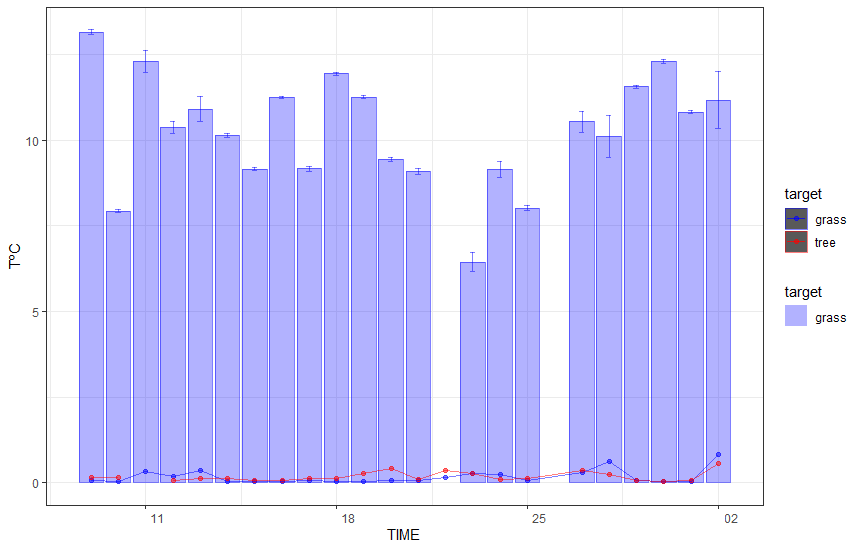
Technically here is your code with corrected synthax error. However I would like to attract your attention to the fact that there is almost no data for tree category and there is no obvious need to plot points and line for 30*sd_NDVI. Probably it would be better to filter the dataset and remove unused geoms. You will also need to adjist legend as well.
You may adjust the desired output according to your needs.
NDVI %>%
mutate(target = as.factor(target)) %>%
ggplot()
geom_bar(aes(x=date, y=TempAmb_Avg, fill = target, color = target),
stat="identity")
geom_point(aes(x=date, y=30*sd_NDVI, color=target),
stat="identity")
geom_line(aes(x=date, y=30*sd_NDVI, color=target),
stat="identity")
geom_errorbar(aes(x = date,
y = TempAmb_Avg,
ymin=TempAmb_Avg-30*sd_NDVI,
ymax=TempAmb_Avg 30*sd_NDVI,
width=.2,,
color = target
),# position = position_dodge(0.05),
stat="identity")
scale_color_manual(values = c(adjustcolor("blue", 0.5), adjustcolor("red", 0.5)), breaks = c("grass", "tree"))
scale_fill_manual(values = c(adjustcolor("blue", 0.3), adjustcolor("red", 0.3)), breaks = c("grass", "tree"))
labs(x="TIME",y="TºC")
theme_bw()
You will get somrthing like this:
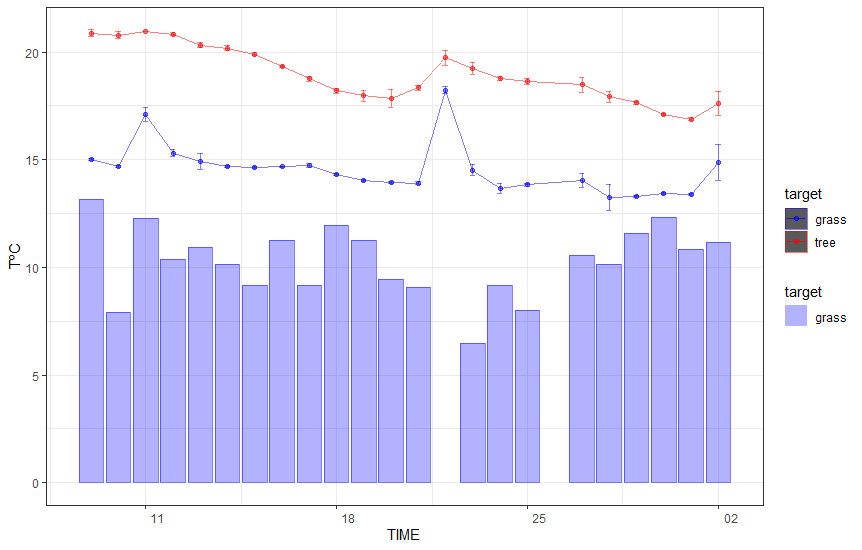
EDITED
upon the edit of the question and comment:
NDVI %>%
mutate(target = as.factor(target)) %>%
ggplot()
geom_bar(aes(x=date, y=TempAmb_Avg, fill = target, color = target),
stat="identity")
geom_point(aes(x=date, y=30*cNDVI , color=target),
#position = position_dodge(),
stat="identity")
geom_line(aes(x=date, y=30*cNDVI, color=target),
#position = position_dodge(),
stat="identity")
geom_errorbar(aes(x = date,
y = cNDVI ,
ymin=30*cNDVI -30*sd_NDVI,
ymax=30*cNDVI 30*sd_NDVI,
width=.2,,
color = target
),
#position = position_dodge(),
stat="identity")
scale_color_manual(values = c(adjustcolor("blue", 0.5), adjustcolor("red", 0.5)), breaks = c("grass", "tree"))
scale_fill_manual(values = c(adjustcolor("blue", 0.3), adjustcolor("red", 0.3)), breaks = c("grass", "tree"))
labs(x="TIME",y="TºC")
theme_bw()