I am struggling with creating multiple ggplots using a loop. I use data in the following format:
a <- c(1,2,3,4)
b <- c(5,6,7,8)
c <- c(9,10,11,12)
d <- c(13,14,15,16)
time <- c(1,2,3,4)
data <- cbind(a,b,c,d,time)
What I want to create is a list of plots that plot one of the letters against the variable time. Which I tried in the following way:
library(ggplot2)
library(gridExtra)
plots <- list()
for (i in 1:4){
plots[[i]] <- ggplot() geom_line(data = data, aes(x = time, y = data[,i]))
}
grid.arrange(plots[[1]], plots[[2]], plots[[3]], plots[[4]])
This results in four times the fourth plot. How do I index this correctly in a way that creates the four intended plots?
CodePudding user response:
(Up front: the reason that your plots are all identical is due to ggplot's "lazy" evaluation of code. See my #2 below, where I identify that the data[,i] is evaluated when you try to plot the data, at which point i is 4, the last pass in the for loop.)
It's generally preferred/recommended to use
data.frames instead of matrices or vectors (as you're doing here). It gives a bit more power and control.data <- data.frame(a,b,c,d,time)Also, I tend to prefer
lapplytofor-loops andlists, for various (some subjective) reasons. Ultimately, the issue you're having is thatggplot2is evaluating the data lazily, soplotsis a list with four plots that make reference toi... and that is realized when you try to plot them all, at which pointiis 4 (from the last pass through the loop). One benefit of usinglapplyis that theireferenced is a local-only (inside of the anon-func) version ofithat is preserved as you would expect.plots <- lapply(names(data)[1:4], function(nm) ggplot(data, aes(x = time, y = .data[[nm]])) geom_line()) gridExtra::grid.arrange(plots[[1]], plots[[2]])I also prefer
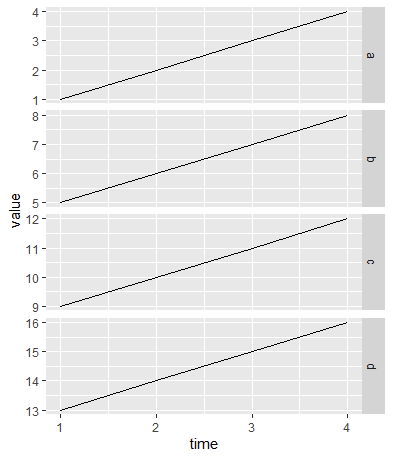
patchworktogridExtra, mostly because it makes more-customized layouts a bit more intuitive, plus adds functionality such as axis-alignment, shared legends, shared titles, etc. (None of those other features are demonstrated here.)library(patchwork) plots[[1]] / plots[[2]] # same plot plots[[1]] plots[[2]] # side-by-side instead of top/bottom (plots[[1]] plots[[2]]) / (plots[[3]] plots[[4]]) # gridUltimately, though, I suggest that facets can be useful and very powerful. For this, we need to melt/pivot the data into a "long format" so that the column names
a-bare actually in one column.reshape2::melt(data, id.vars = "time") |> ggplot(aes(time, value)) geom_line() facet_grid(variable ~ ., scales = "free_y")I assumed the preference for independent (free) y-scales, ergo the
scales="free_y". Try it without if you want to see the options. (There are alsoscales="free_x"andscales="free"(both).)To see what I mean by "long" format:
reshape2::melt(data, id.vars = "time") # time variable value # 1 1 a 1 # 2 2 a 2 # 3 3 a 3 # 4 4 a 4 # 5 1 b 5 # 6 2 b 6 # 7 3 b 7 # 8 4 b 8 # 9 1 c 9 # 10 2 c 10 # 11 3 c 11 # 12 4 c 12 # 13 1 d 13 # 14 2 d 14 # 15 3 d 15 # 16 4 d 16This can also be done with
tidyr::pivot_longer(data, -time), albeit thevariablename is nowname. For this use, there is no advantage toreshape2::meltortidyr::pivot_longer; there are opportunities for significantly more complex pivoting in the latter, not relevant with this data.
Data
data <- structure(list(a = c(1, 2, 3, 4), b = c(5, 6, 7, 8), c = c(9, 10, 11, 12), d = c(13, 14, 15, 16), time = c(1, 2, 3, 4)), class = "data.frame", row.names = c(NA, -4L))