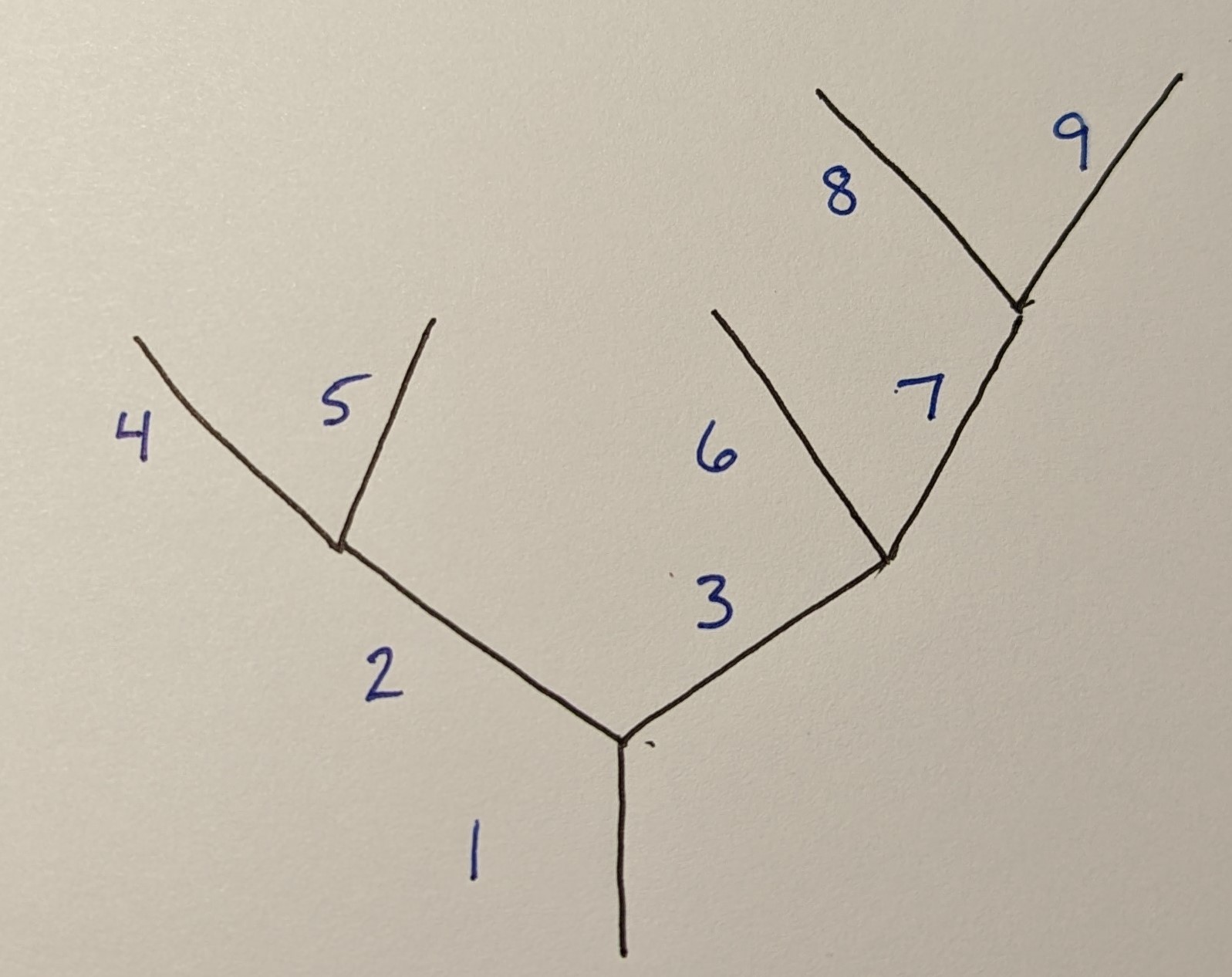
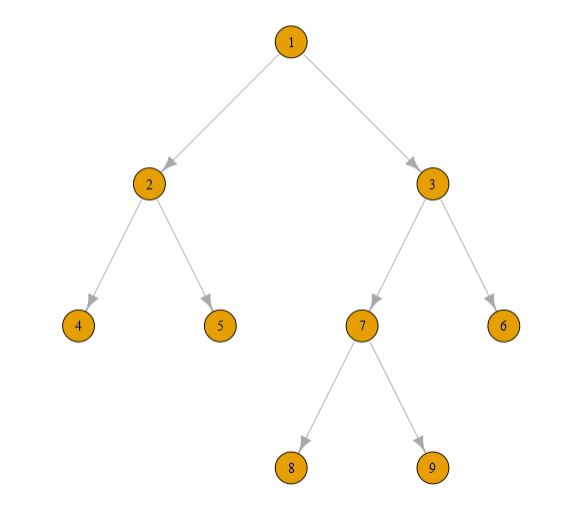
I have a table that defines a binary tree with thousands of individual links. The table has arbitrarily defined, numeric link names, arbitrarily defined parent links (PL) (where -1 indicates no parent) and there is never 1 parent, and the length of each link. I would like to construct a table that has each link and its total distance from the base of link 1.
For the above network I would have this data.frame though in practice there is no pattern to the naming of the links:
test_data <- data.frame(Link = seq(1, 9, 1),
PL1 = c(2, 4, 6, -1, -1, -1, 8, -1, -1),
PL2 = c(3, 5, 7, -1, -1, -1, 9, -1, -1),
Len = c(15, 17, 81, 9, 42, 7, 13, 64, 36))
And I would like to have a table like this:
result <- data.frame(Link = seq(1, 9, 1),
Len = c(15, 15 17, 15 81, 15 17 9, 15 17 42, 15 81 7, 15 81 13, 15 81 13 64, 15 81 13 36))
where, for example, the length of Link7 (L7) = L1 L3 L7. The above code shows this work but I would like the table to contain the total sum. Thanks!
CodePudding user response:
Here is one way using the igraph package. It has a plethora of functions so there might be a more direct way but this gets you to your desired result:
library(igraph)
library(tidyverse)
plot_dat <- test_data %>%
pivot_longer(-c(Link, Len)) %>%
select(Link, value) %>%
filter(value != -1) %>%
graph_from_data_frame()
plot_dat %>%
plot(layout = layout_as_tree(.))
n <- neighborhood(plot_dat, mode = "in")
all_simple_paths(plot_dat, from = as_ids(n[lengths(n) == 1][[1]])) %>%
map(~ as.numeric(as_ids(.x))) %>%
enframe(value = "path") %>%
mutate(Link = map_dbl(path, last)) %>%
left_join(test_data, ., by = "Link") %>%
mutate(Len_total = replace(map_dbl(path, ~ sum(Len[match(.x, Link)])), 1, Len[1])) %>%
select(Link, Len = Len_total)
Link Len
1 1 15
2 2 32
3 3 96
4 4 41
5 5 74
6 6 103
7 7 109
8 8 173
9 9 145