import pandas as pd
data = {'A': ['123','456','789'], 'B': ['D1','D4','D7'], 'C':['D2','D5','D8'], 'D':['D3','D6','D9']}
df1 = pd.DataFrame(data)
data2 = {'A': ['123','789','111','222'], 'B': ['D1','D7','D11','D14'], 'C':['D10','D8','D12','D15'], 'D':['D3','D9','D13','16']}
df2 = pd.DataFrame(data2)
The primary key link between the 2 frames is column 'A'.
Output desired -
- df1 - rows which are not present in df2 ('A' = 456)
- df2 - rows which are not present in df1 ('A' = 111 and 'A' = 222)
- common rows with differences - ('A' = 123 - column 'C' = D2 vs D10
CodePudding user response:
Q1: df1 - rows which are not present in df2:
q1=df1[~df1['A'].isin(df2['A'])]
'''
A B C D
1 456 D4 D5 D6
'''
Q2: df2 - rows which are not present in df1:
q2=df2[~df2['A'].isin(df1['A'])]
'''
A B C D
2 111 D11 D12 D13
3 222 D14 D15 16
'''
Q3: Common rows with differences:
q3=pd.concat([df1[df1['A'].isin(df2['A'])],df2[df2['A'].isin(df1['A'])]]).drop_duplicates().groupby('A').agg(list)
mask=q3.applymap(lambda x: pd.Series(x).is_unique if len(x) > 1 else False)
q3=q3[mask].stack().reset_index()
'''
A level_1 0
0 123 C [D2, D10]
'''
CodePudding user response:
I have something similar to @Bushmaster's answer for the first two questions. But a slightly different approach to show the differences among the rows where A is common.
df1x2 = df1.loc[~df1['A'].isin(df2['A'])]
df2x1 = df2.loc[~df2['A'].isin(df1['A'])]
dfcom = df1.merge(df2, on='A', suffixes=['_1', '_2'])
Then, to highlight the differences within each row of dfcom:
def show_diffs(s):
a = {k[:-2]: v for k, v in s.items() if k.endswith('_1')}
b = {k[:-2]: v for k, v in s.items() if k.endswith('_2')}
return {k: (a[k], b[k]) for k in a if a[k] != b[k]}
>>> dfcom.assign(diffs=dfcom.apply(show_diffs, axis=1))
A B_1 C_1 D_1 B_2 C_2 D_2 diffs
0 123 D1 D2 D3 D1 D10 D3 {'C': ('D2', 'D10')}
1 789 D7 D8 D9 D7 D8 D9 {}
# or, just the differences
>>> dfcom[['A']].assign(diffs=dfcom.apply(show_diffs, axis=1))
A diffs
0 123 {'C': ('D2', 'D10')}
1 789 {}
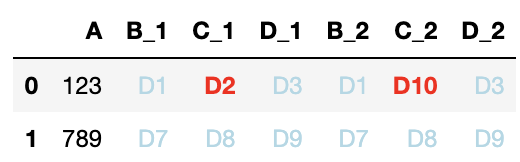
Another possibility, if the common dfcom is not too large, is to present the differences visually:
def highlight(df):
cols_1 = [k for k in df.columns if k.endswith('_1')]
cols_2 = [k for k in df.columns if k.endswith('_2')]
a = df[cols_1].set_axis([k[:-2] for k in cols_1], axis=1)
b = df[cols_2].set_axis([k[:-2] for k in cols_2], axis=1)
h = pd.concat([
# just in case the columns are somehow in a different order
(a != b[a.columns]).set_axis(cols_1, axis=1),
(b != a[b.columns]).set_axis(cols_2, axis=1),
], axis=1).applymap(
lambda v: 'color:red; font-weight:bold' if v else 'color:lightblue')
return h
dfcom.style.apply(highlight, axis=None)
CodePudding user response:
Here's a way to do what you've asked by:
- changing the index to
A - pre-calculating subsets of common and distinct values of
A - reading off the appropriate rows by index (i.e., by value of
A) for non-common rows indf1anddf2 - creating a mask for common rows flagging non-matching values and using this to construct
V1 vs V2style strings in positions that have differing values, with like values marked with NaN.
# pre-calculation of partitioning of `A` values, to avoid duplicate operations
a1, a2 = df1.set_index('A'), df2.set_index('A')
s1, s2 = set(a1.index), set(a2.index)
sCommon = s1 & s2
lCommon = list(sCommon)
# calculation of outputs (rows only in df1, rows only in df2, and common rows)
out1, out2 = a1.loc[list(s1 - sCommon),], a2.loc[list(s2 - sCommon),]
mask = a1.loc[lCommon,] != a2.loc[lCommon,]
out3 = (a1.loc[lCommon,].astype(str)[mask] ' vs ' a2.loc[lCommon,].astype(str)[mask])
Output:
out1
B C D
A
456 D4 D5 D6
out2
B C D
A
222 D14 D15 16
111 D11 D12 D13
out3
B C D
A
123 NaN D2 vs D10 NaN
789 NaN NaN NaN
