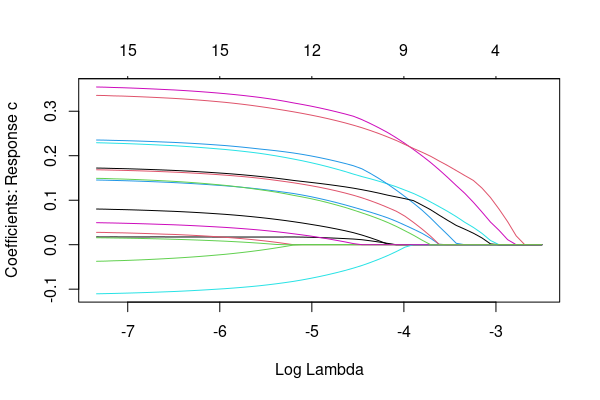
I'm attemtping to select a single grpah from multiple plot output with "glmnet" function (especially elastic net) in R.
I'm currently dealing with multinomial case; the response variable has 3 or more categories. So I fit the model with explanatory variable X and the response variable y with 3 types of categories. Assume that we already have alpha and lambda values from cross validation using cv.glmnet. I didn't use the argument type.multinomial = "grouped".
fit <- glmnet(X, y, alpha = alpha, lambda = lambda, standardize = TRUE, family = "multinomial")
plot(fit, xvar = "lambda")
fit is then the elastic net model. I know that plot(fit, xvar = "lambda") shows the coefficients path something like this:
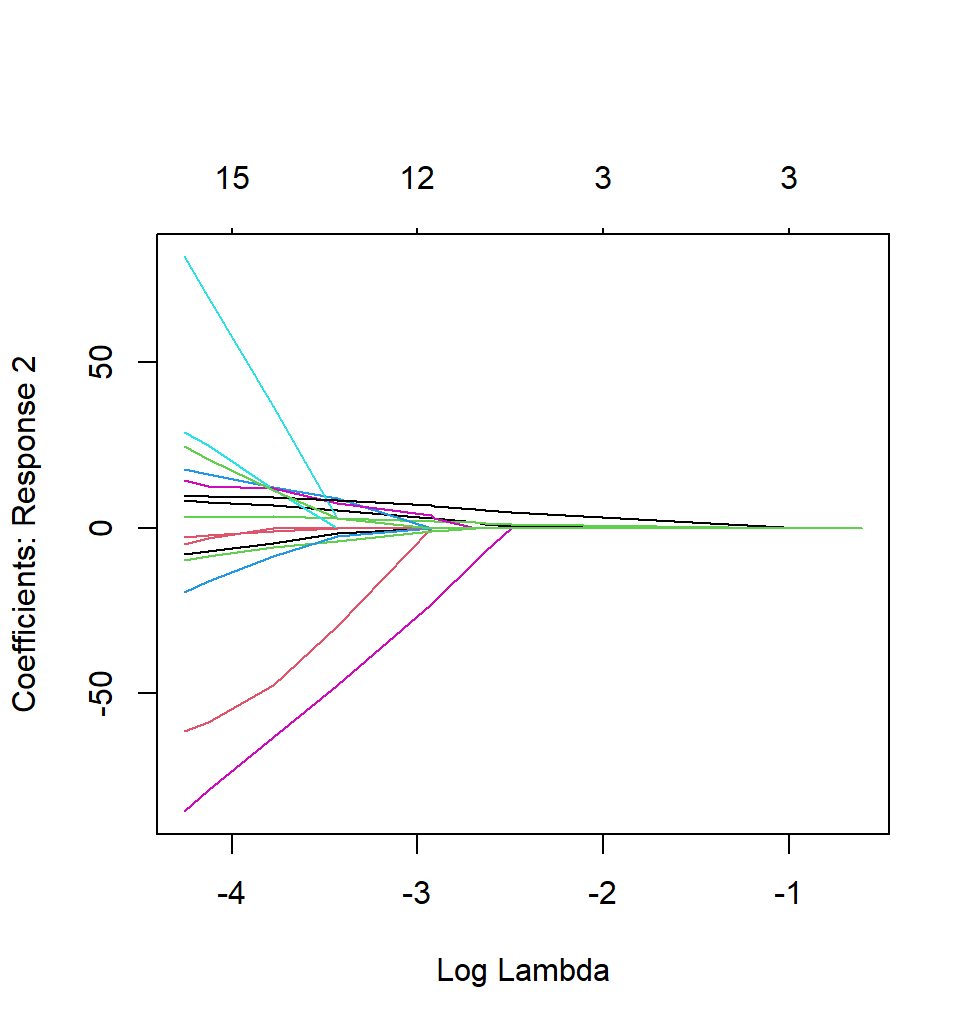
However, when it is multinomial case, R generates multiple coefficient plots at the same time: for example when y has three categories, then R shows 3 coefficient plots. But I need to select only one graph out of them. I tried to find a way in google, but failed. Is there any way that I can try?
CodePudding user response:
We could modify the glmnet:::plot.multnet method for this and add a which= option similar to that in stats:::plot.lm.
plot.multnet <- function(x, xvar=c("norm", "lambda", "dev"), label=FALSE,
type.coef=c("coef", "2norm"), which=x$classnames, ...) {
xvar <- match.arg(xvar)
type.coef <- match.arg(type.coef)
if (!all(which %in% x$classnames)) {
warning(sprintf('`which=%s` not in classnames, defaulting to all', which))
which <- x$classnames
}
beta <- x$beta
if (xvar == "norm") {
cnorm1 <- function(beta) {
whichnz <- nonzeroCoef(beta)
beta <- as.matrix(beta[whichnz, ])
apply(abs(beta), 2, sum)
}
norm <- apply(sapply(x$beta, cnorm1), 1, sum)
} else {
norm <- NULL
}
dfmat <- x$dfmat
if (type.coef == "coef") {
ncl <- nrow(dfmat)
clnames <- rownames(dfmat)
lapply(which, function(z) glmnet:::plotCoef(beta[[z]], norm, x$lambda, dfmat[z, ], x$dev.ratio,
label=label, xvar=xvar, ylab=paste("Coefficients: Response",
grep(z, clnames, value=TRUE))))
} else {
dfseq <- round(apply(dfmat, 2, mean), 1)
glmnet:::plotCoef(coefnorm(beta, 2), norm, x$lambda, dfseq, x$dev.ratio,
label=label, xvar=xvar, ylab="Coefficient 2Norms",
...)
}
}
Usage
fit1 <- glmnet(x, y, family="multinomial")
plot(fit1, xvar='lambda', which='c')
Throws a warning if incorrectly specified and plots all levels.
plot(fit1, xvar='lambda', which=1)
# Warning message:
# In plot.multnet(fit1, xvar = "lambda", which = 1) :
# `which=1` not in classnames, defaulting to all
Data:
set.seed(42)
x <- matrix(rnorm(100 * 20), 100, 20)
y <- sample(letters[1:3], 100, replace=TRUE)
CodePudding user response:
Here is a way, hacking the code of plot.multnet, like in jay.sf's answer. The hacking is different, though. It doesn't throw a warning when asked to plot all levels. See the code comments on what is done.
The data set and fit example is taken from the documentation of plot.multnet, see, for instance, here.
library(glmnet)
#> Loading required package: Matrix
#> Loaded glmnet 4.1-4
plot.multnet <-
function (x, xvar = c("norm", "lambda", "dev"), label = FALSE,
type.coef = c("coef", "2norm"), which = NULL, ...)
{
xvar = match.arg(xvar)
type.coef = match.arg(type.coef)
beta = x$beta
if (xvar == "norm") {
cnorm1 = function(beta) {
which = glmnet:::nonzeroCoef(beta)
beta = as.matrix(beta[which, ])
apply(abs(beta), 2, sum)
}
norm = apply(sapply(x$beta, cnorm1), 1, sum)
}
else norm = NULL
dfmat = x$dfmat
if (type.coef == "coef") {
ncl = nrow(dfmat)
clnames = rownames(dfmat)
#
if(is.null(which)) { # if all response values are to be plotted
seq_ncl <- seq(ncl) # use the package's loop control variable
} else {
seq_ncl <- which # if not, only the wanted values
}
#
for (i in seq_ncl) {
glmnet:::plotCoef(beta[[i]], norm, x$lambda, dfmat[i, ], x$dev.ratio,
label = label, xvar = xvar, ylab = paste("Coefficients: Response",
clnames[i]), ...)
}
}
else {
dfseq = round(apply(dfmat, 2, mean), 1)
glmnet:::plotCoef(coefnorm(beta, 2), norm, x$lambda, dfseq, x$dev.ratio,
label = label, xvar = xvar, ylab = "Coefficient 2Norms",
...)
}
}
set.seed(2022)
x <- matrix(rnorm(100*20), 100, 20)
g4 <- sample(4, 100, replace = TRUE)
fit3 <- glmnet(x, g4, family = "multinomial")
op <- par(mfrow = c(2, 2))
plot(fit3)
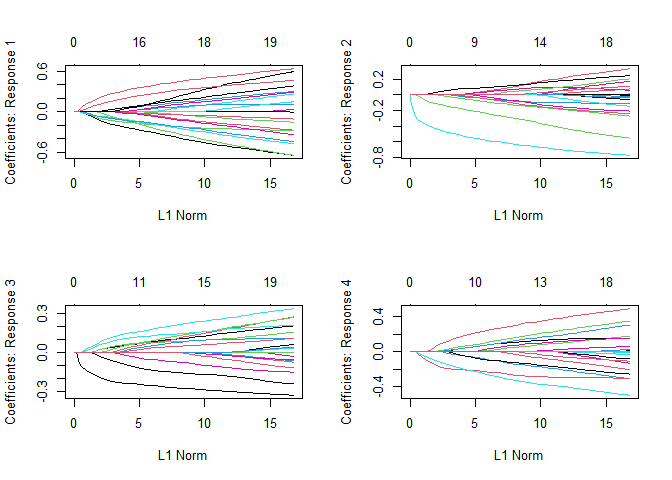
par(op)
plot(fit3, which = 2)
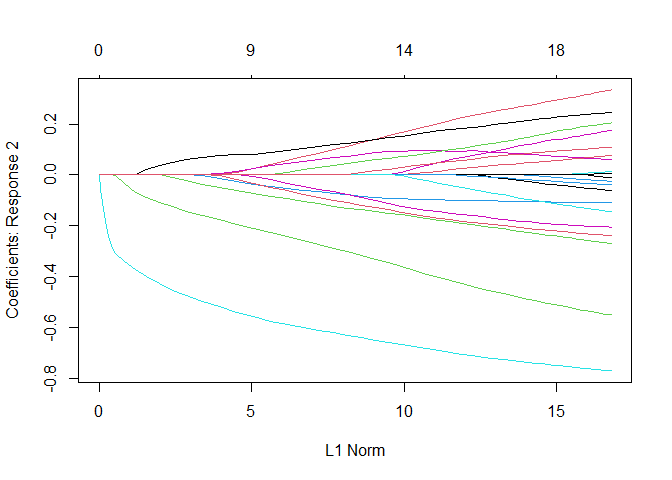
Created on 2022-12-19 with reprex v2.0.2