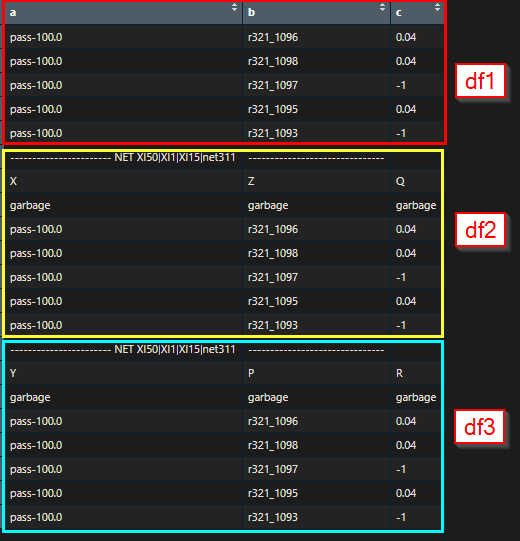
I am trying to split a very big dataframe into multiple dataframes where column value starts with "-----..." and until I reach "-----..."
I have a big dataframe (made by reading a text file and later prepared into a df) containing more than 1.9m rows,
a = c("pass-100.0","pass-100.0","pass-100.0","pass-100.0","pass-100.0","----------------------- NET XI50|XI1|XI15|net311","X","garbage","pass-100.0","pass-100.0","pass-100.0","pass-100.0","pass-100.0","----------------------- NET XI50|XI1|XI15|net311","Y","garbage","pass-100.0","pass-100.0","pass-100.0","pass-100.0","pass-100.0")
b = c("r321_1096","r321_1098","r321_1097","r321_1095","r321_1093","-------------------------------","Z","garbage","r321_1096","r321_1098","r321_1097","r321_1095","r321_1093","-------------------------------","P","garbage","r321_1096","r321_1098","r321_1097","r321_1095","r321_1093")
c = c("0.04","0.04","-1","0.04","-1","","Q","garbage","0.04","0.04","-1","0.04","-1","","R","garbage","0.04","0.04","-1","0.04","-1")
d = c("0.32","0.32","0","0.32","0","","S","garbage","0.32","0.32","0","0.32","0","","T","garbage","0.32","0.32","0","0.32","0")
df = data.frame(a, b, c, d)
I want to split the df into smaller dataframes where column 'a' starts with "----------------------- NET XI50|XI1|XI15|net311" and the dataframe ends when it encounters "----------------------- NET XI50|XI1|XI15|net311" for the second time.
I want to keep the column names as a, b and c for all the new dataframes and remove the row containing "garbage" values in the smaller dataframes. Later, I plan on doing some common calculations on these dataframes. But I cant think of a way to split the big dataframe into usable ones. Any idea how I should proceed?
CodePudding user response:
You can use the split function to do this. It relies on making a factor corresponding to each group, which you can get using cumsum to count the number of rows that have matched to ---- in column A. The result is a list of data frames (I've also filtered out the 'garbage' row):
split(df[df$a!="garbage",] , cumsum(grepl("----",df[df$a!="garbage","a"])))
$`0`
a b c d
1 pass-100.0 r321_1096 0.04 0.32
2 pass-100.0 r321_1098 0.04 0.32
3 pass-100.0 r321_1097 -1 0
4 pass-100.0 r321_1095 0.04 0.32
5 pass-100.0 r321_1093 -1 0
$`1`
a b c d
6 ----------------------- NET XI50|XI1|XI15|net311 -------------------------------
7 X Z Q S
9 pass-100.0 r321_1096 0.04 0.32
10 pass-100.0 r321_1098 0.04 0.32
11 pass-100.0 r321_1097 -1 0
12 pass-100.0 r321_1095 0.04 0.32
13 pass-100.0 r321_1093 -1 0
$`2`
a b c d
14 ----------------------- NET XI50|XI1|XI15|net311 -------------------------------
15 Y P R T
17 pass-100.0 r321_1096 0.04 0.32
18 pass-100.0 r321_1098 0.04 0.32
19 pass-100.0 r321_1097 -1 0
20 pass-100.0 r321_1095 0.04 0.32
21 pass-100.0 r321_1093 -1 0
CodePudding user response:
Or a dplyr equivalent using (the experimental, but handy) group_split.
I would, however, recommend that you do this on the txt before you create the data frames: It's (probably) easier, safer, and faster for 1.9m rows.
library(dplyr)
library(stringr)
df |>
group_by(group = cumsum(str_detect(a,
fixed("----------------------- NET XI50|XI1|XI15|net311")))) |> # Fixed is faster but approximate
filter(between(row_number(), 4, n()) & group > 0 | group == 0) |>
group_split(.keep = FALSE)
Output:
[[1]]
# A tibble: 5 × 4
a b c d
<chr> <chr> <chr> <chr>
1 pass-100.0 r321_1096 0.04 0.32
2 pass-100.0 r321_1098 0.04 0.32
3 pass-100.0 r321_1097 -1 0
4 pass-100.0 r321_1095 0.04 0.32
5 pass-100.0 r321_1093 -1 0
[[2]]
# A tibble: 5 × 4
a b c d
<chr> <chr> <chr> <chr>
1 pass-100.0 r321_1096 0.04 0.32
2 pass-100.0 r321_1098 0.04 0.32
3 pass-100.0 r321_1097 -1 0
4 pass-100.0 r321_1095 0.04 0.32
5 pass-100.0 r321_1093 -1 0
[[3]]
# A tibble: 5 × 4
a b c d
<chr> <chr> <chr> <chr>
1 pass-100.0 r321_1096 0.04 0.32
2 pass-100.0 r321_1098 0.04 0.32
3 pass-100.0 r321_1097 -1 0
4 pass-100.0 r321_1095 0.04 0.32
5 pass-100.0 r321_1093 -1 0
CodePudding user response:
I'd suggest doing something like this:
df |>
mutate(group = cumsum(a |> str_starts("---"))) |>
nest(data = -group) |>
rowwise(group) |>
summarise(
data = list(
if (group == 0) data else slice_tail(data, n = -3)
)
)
#> # A tibble: 3 × 2
#> group data
#> <int> <list>
#> 1 0 <tibble [5 × 4]>
#> 2 1 <tibble [5 × 4]>
#> 3 2 <tibble [5 × 4]>
Explanation: First, we create a column that indicates when each new chunk starts
df |>
mutate(
group = cumsum(a |> str_starts("----"))
)
#> # A tibble: 21 × 5
#> a b c d group
#> <chr> <chr> <chr> <chr> <int>
#> 1 pass-100.0 r321_1096 "0.0… "0.3… 0
#> 2 pass-100.0 r321_1098 "0.0… "0.3… 0
#> 3 pass-100.0 r321_1097 "-1" "0" 0
#> 4 pass-100.0 r321_1095 "0.0… "0.3… 0
#> 5 pass-100.0 r321_1093 "-1" "0" 0
#> 6 ----------------------- NET XI50|XI1|XI15|net311 ---------… "" "" 1
#> 7 X Z "Q" "S" 1
#> 8 garbage garbage "gar… "gar… 1
#> 9 pass-100.0 r321_1096 "0.0… "0.3… 1
#> 10 pass-100.0 r321_1098 "0.0… "0.3… 1
#> # … with 11 more rows
and now you could nest the groups into separate dataframes
df |>
mutate(
group = cumsum(a |> str_starts("----"))
) |>
nest(data = -group)
#> # A tibble: 3 × 2
#> group data
#> <int> <list>
#> 1 0 <tibble [5 × 4]>
#> 2 1 <tibble [8 × 4]>
#> 3 2 <tibble [8 × 4]>
which then can be processed using a combination of rowwise and summarise
df |>
mutate(
group = cumsum(a |> str_starts("----"))
) |>
nest(data = -group) |>
rowwise(group) |>
summarise(
data = list(
if (group == 0) data else slice_tail(data, n = -3)
)
)
#> # A tibble: 3 × 2
#> group data
#> <int> <list>
#> 1 0 <tibble [5 × 4]>
#> 2 1 <tibble [5 × 4]>
#> 3 2 <tibble [5 × 4]>