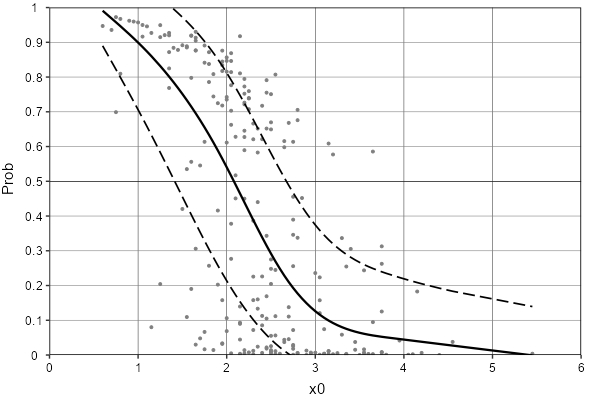
I'd' like to model the 25th, 50th and 75th quantile regression curves (q25, q50, q75) for 241 values of probability ('prob') depending on x0.
For that purpose, I used the qgamV package as follows. However, this approach led to some q25, q50, q75 values <0 and >1, which is not expected for probabilities. Graphically, one would expect the q25 and q75 regression curves to approach the 'prob' limits 0 and 1 in a more tangential way (see below).
How to model these quantiles curves as best as possible, knowing that they represent probabilities?
Thanks for help.
Initial dataframe (df0):
df0 <- structure(list(x0 = c(2.65, 3.1, 2.15, 2.45, 2.9, 1.55, 2.05,
2.75, 2, 2.45, 4.05, 1.95, 3.35, 2.15, 2.5, 1.75, 1.6, 2.3, 3.35,
3.55, 2.1, 3.15, 2.5, 1.05, 2.3, 2.3, 2.95, 0.8, 1.75, 2.95,
2.55, 1.65, 2.4, 2.8, 2.2, 3.45, 2.15, 2.9, 1.7, 2.7, 2.05, 2.75,
2.35, 3.75, 2.2, 1.1, 2.35, 2.5, 3.05, 1, 4.4, 1.3, 2.2, 2.5,
1.35, 1.95, 1.95, 5.45, 2, 1.65, 2.7, 2, 1.5, 1.05, 4.15, 2.15,
1.9, 1.85, 4.2, 2.2, 3.35, 1.55, 1.95, 2.3, 1.9, 3.45, 2.2, 3.55,
1.4, 2.5, 2.35, 2.5, 2.4, 3.35, 2, 2.6, 3.05, 2.75, 1.6, 1.65,
2.45, 1.55, 1.65, 2.25, 0.9, 2.4, 2.2, 2, 1.65, 1.35, 1.95, 2.5,
1.6, 1.25, 3.8, 2.25, 2.85, 1.45, 2.4, 2.8, 3.75, 3.05, 1.8,
1.25, 1.55, 2, 2.55, 2.75, 3.55, 2.2, 2.1, 3.55, 3.65, 2.3, 1.25,
2.45, 2.2, 1.95, 1.65, 0.7, 2, 1.5, 2.8, 3.4, 3.95, 2.55, 2.45,
2.65, 1.75, 1.7, 2.5, 2.05, 2.75, 2.05, 3, 2.25, 3.6, 2.35, 3.25,
1.6, 3.3, 2.05, 1.95, 2.15, 2.3, 4.1, 2.45, 1.6, 2.3, 0.6, 2.35,
2.45, 1.9, 2.5, 1.35, 3.2, 2.25, 1.65, 2.75, 1.8, 3, 0.95, 2.7,
2.15, 3.75, 2.5, 1.95, 2.7, 3.75, 2.4, 2.4, 3.05, 1.8, 3.6, 2.05,
2.75, 2.15, 1.35, 3.15, 2.25, 3.1, 2, 2.35, 3.3, 2.05, 0.75,
2.55, 2.2, 3.15, 3.1, 1.75, 3.2, 3.15, 2.8, 2.5, 1.8, 2.2, 1.85,
3.35, 1.35, 2.75, 1.85, 2.8, 2.65, 3.15, 1.15, 2.5, 3.75, 2.75,
4.55, 2.3, 2.65, 3.1, 3.65, 0.8, 2.45, 3.25, 3.65, 3.75, 1.75,
2.55, 1.15, 2.05, 2.05, 3.5, 0.75, 2.55, 2.2, 2.1, 2.15, 2.75
), prob = c(0.043824528975438, 0.0743831343145038, 0.0444802301649798,
0.0184204002808217, 0.012747152819121, 0.109320069103749, 0.868637913750677,
0.389605665620339, 0.846536935687218, 0.104932383728924, 0.000796924809569913,
0.844673988202945, 0.00120791067227541, 0.91751061807481, 0.0140582427585067,
0.61360854266884, 0.55603090737844, 0.0121424615930165, 0.000392412410090414,
0.00731972612592678, 0.450730636411052, 0.0111896050578429, 0.0552971757296455,
0.949825608148576, 0.00216318997302124, 0.620876890784462, 0.00434032271743834,
0.809464444601336, 0.890796570916792, 0.0070834616944228, 0.0563350845256127,
0.913156468748195, 0.00605085671490011, 0.00585882020388307,
0.0139577135093548, 0.0151356267602558, 0.00357231467872644,
0.000268107682417655, 0.047883018897558, 0.137688264298974, 0.846219411361109,
0.455395192661041, 0.440089914302649, 0.312776912863294, 0.721283899836456,
0.945808616162847, 0.160122538485323, 0.274966581834218, 0.223500907500226,
0.957169102670141, 3.29173412975754e-05, 0.920710197397359, 0.752055893010363,
0.204573327883464, 0.824869881489217, 0.0336636091577387, 0.834235793851965,
0.00377210373002217, 0.611370672834389, 0.876156793482752, 0.04563653558985,
0.742493995255321, 0.42035122692417, 0.916359628728296, 0.182755925347698,
0.139504394672643, 0.415836463269909, 0.0143112277191436, 0.00611022961831899,
0.794529254262237, 0.000295836911230635, 0.88504245090271, 0.0320097205131667,
0.386424550101868, 0.724747784339428, 0.0374198694261709, 0.772894216412908,
0.243626917726206, 0.884082536765856, 0.649357153222083, 0.651665475576256,
0.248153637183556, 0.621116026311962, 0.254679380328883, 0.815492354289526,
0.00384382735772974, 0.00098493832845314, 0.0289740210412282,
0.919537164719931, 0.029914235716672, 0.791051705450356, 0.535062926433525,
0.930153425256182, 0.739648381556949, 0.962078822556967, 0.717404075711021,
0.00426200695619151, 0.0688025266083751, 0.30592683399928, 0.76857384388609,
0.817428136470741, 0.0101583095649087, 0.190150584186769, 0.949353043876038,
0.000942385744019884, 0.00752842476126574, 0.451811230189468,
0.878142444707428, 0.085390660867941, 0.705492062082986, 0.00776625091631656,
0.120499683875168, 0.871558791341612, 0.204175216963286, 0.88865934672351,
0.735067195665991, 0.111767657566763, 0.0718305257427526, 0.001998068594943,
0.726375812318976, 0.628064249939129, 0.0163105011142307, 0.585565544471761,
0.225632568540361, 0.914834452659588, 0.755043268549628, 0.44993311080756,
0.876058522964169, 0.876909380258345, 0.935545943209396, 0.856566304797687,
0.891579321327903, 0.67586664661773, 0.305274362445618, 0.0416387565225755,
0.244843991055886, 0.651782914419153, 0.615583040148267, 0.0164959661557421,
0.545479687527543, 0.0254178939123714, 0.00480000384583597, 0.0256296636591875,
0.776444262284288, 0.00686736233661002, 0.738267311816833, 0.00284628668554737,
0.0240371572079387, 0.00549270830047392, 0.91880163437759, 0.336534358175717,
0.276841848679916, 0.718008645244615, 0.0897424253787563, 0.0719730540202573,
0.00215797941000608, 0.0219160132143199, 0.797680147185277, 0.66612383359622,
0.946965411044528, 0.133399527090937, 0.343056247984854, 0.202570454449074,
0.00349712323805031, 0.919979740593237, 0.577123238372546, 0.759418264563034,
0.904569159000302, 0.0179587619909363, 0.785657258439329, 0.235867625712547,
0.959688292861383, 0.668060191654474, 0.0014774986557077, 0.00831528722028647,
0.669655207261098, 0.157824457113222, 0.110637023939517, 0.262525772704882,
0.112654002253028, 0.22606090266161, 0.157513622503487, 0.25688454756606,
0.00201570863346944, 0.70318409224183, 0.25568985167711, 0.810637054896326,
0.92708070974999, 0.608664352336801, 0.707490903842404, 0.00094520948858089,
0.106177223644193, 0.582785205597368, 0.0585327568963445, 0.377814739935042,
0.972447647118833, 0.0111118791692372, 0.58947840090326, 0.0111189166236961,
0.00317374095338712, 0.0664218007312096, 0.00227258301798719,
0.00198861129291917, 0.337443337988163, 0.750708293355867, 0.837530172974158,
0.627428065068903, 0.744110974625108, 0.00320417425932798, 0.871800026765784,
0.613647987816266, 0.808457030433619, 0.00486495461698562, 0.597950577021363,
0.000885253981642748, 0.0800527366346806, 0.00951706823839207,
0.125222576598629, 0.346018567766834, 0.0376933970313487, 0.157903106929268,
0.0371982251307384, 0.00407175432189843, 0.0946588147179984,
0.967274516618573, 0.169109953293894, 0.00124072042059317, 0.00259042255361196,
0.000400511359506596, 0.841289470209085, 0.807106898740506, 0.926962245924993,
0.814160745645036, 0.662558468801531, 0.000288068688170646, 0.698932091902567,
0.00242011818508616, 0.645573844423654, 0.517121859568318, 0.0931231998319089,
0.000877774529895907)), row.names = c(NA, -241L), class = "data.frame")
Quantiles regressions and plot:
library(mgcViz)
library(qgam)
library(ggplot2)
# Quantile regressions
q50 <- qgamV(prob ~ s(x0, bs="cr", k=10), data = df0, qu = 0.5)
q25 <- qgamV(prob ~ s(x0, bs="cr", k=10), data = df0, qu = 0.25)
q75 <- qgamV(prob ~ s(x0, bs="cr", k=10), data = df0, qu = 0.75)
# New dataframe including fitted quantile values
df1 <- df0
df1$q50 <- q50[["fitted.values"]]
df1$q25 <- q25[["fitted.values"]]
df1$q75 <- q75[["fitted.values"]]
# Plot
x_brk <- seq(0, 6, 1); x_lab <- seq(0, 6, 1)
y_brk <- seq(0, 1, 0.1); y_lab <- seq(0, 1, 0.1)
ggplot(df1, aes(x = x0, y = prob))
scale_x_continuous(limits=c(0, 20), expand=c(0, 0), breaks=x_brk, labels=x_lab)
scale_y_continuous(limits=c(-1, 2),expand=c(0, 0), breaks=y_brk, labels=y_lab)
geom_vline(xintercept=x_brk, colour="grey25", size=0.2)
geom_hline(yintercept=y_brk, colour="grey50", size=0.2)
geom_hline(yintercept=0.5, linetype="solid", color = "black", size=0.2)
geom_point(data = df1, aes(x = x0, y = prob), colour = "grey50", size=0.75, inherit.aes = TRUE)
xlab(~paste("x0"))
ylab(~paste("Prob"))
theme(plot.title = element_blank())
theme(plot.margin=unit(c(0.2,0.5,0.01,0.3),"cm"))
theme(axis.text.x=element_text(colour="black", size=9.5, margin=margin(b=10),vjust=-1))
theme(axis.text.y=element_text(colour="black", size=9.5,hjust=0.5))
theme(axis.title.x=element_text(colour="black", size=11.5, margin=margin(b=2), vjust=1))
theme(axis.title.y=element_text(colour="black", size=11.5, margin=margin(b=2), vjust=4))
theme(panel.background=element_rect(fill="white"), panel.border = element_rect(colour = "black", fill=NA))
geom_line(aes(x=x0, y = q50), data=df1, colour="black",size=0.8, inherit.aes = TRUE)
geom_line(aes(x=x0, y = q25), data=df1, colour="black",size=0.6, linetype = "longdash")
geom_line(aes(x=x0, y = q75), data=df1, colour="black",size=0.6, linetype = "longdash")
coord_cartesian(xlim = c(0, 6), ylim = c(0, 1))
Continuation of the solution proposed by user2974951:
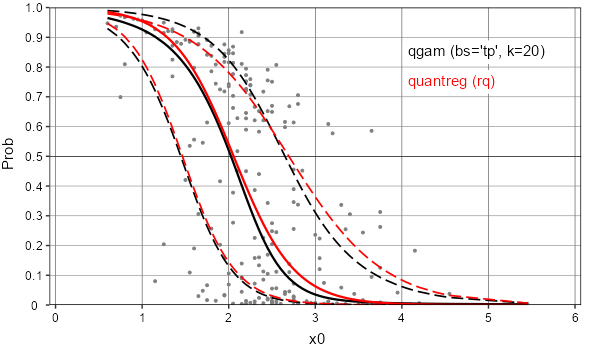
Given the non-normal distribution of Prob, I think better to use qgam rather than quantreg, by taking inspiration from user2974951's solution.
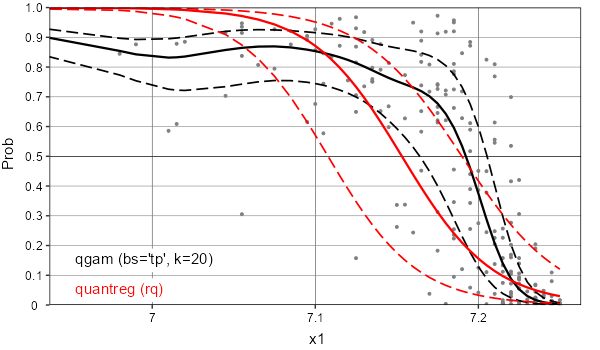
The difference between these 2 quantile regression approaches is very slight on example x0, but much more obvious with another predictor x1:
Example x0:
Example x1:
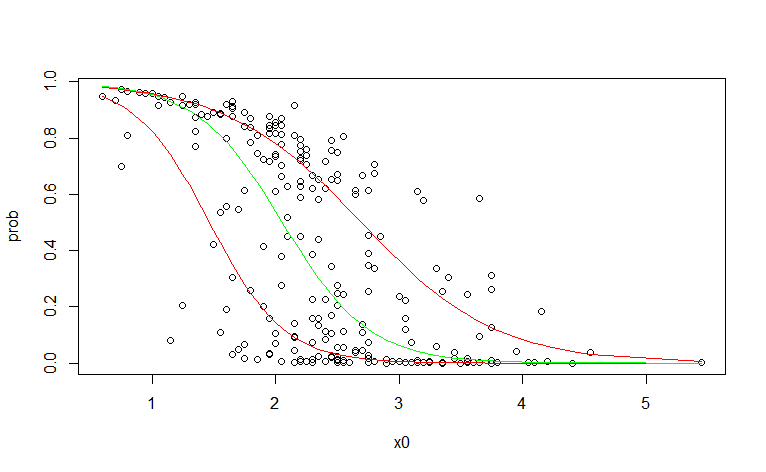
CodePudding user response:
You can use the logit transform and then use regular quantile regresion
library(quantreg)
df0 <- df0[order(df0$x0), ] # ordering just for easier visualization
df0$probL <- log(df0$prob/(1 - df0$prob))
t <- c(0.25, 0.5, 0.75)
mod <- lapply(t, function(x){rq(probL ~ x0, data=df0, tau=x)})
names(mod) <- paste0("Q_", t)
pre <- as.data.frame(do.call(cbind, lapply(mod, function(x){1/(1 exp(-predict(x)))})))
plot(prob ~ x0, data=df0)
lines(pre$Q_0.25 ~ df0$x0, col="red")
lines(pre$Q_0.5 ~ df0$x0, col="green")
lines(pre$Q_0.75 ~ df0$x0, col="red")