I'm able to calculate a rolling correlation coefficient for a 1D-array (data against [0, 1, 2, 3, 4]) using a loop.
I'm looking for a smarter solution using numpy (not pandas).
Here is my current code:
import numpy as np
data = np.array([10,5,8,9,15,22,26,11,15,16,18,7,4,8,-2,-3,-4,-6,-2,0,10,0,5,8])
x = np.zeros_like(data).astype('float32')
length = 5
for i in range(length, data.shape[0]):
x[i] = np.corrcoef(data[i - length:i], np.arange(length))[0, 1]
print(x)
x gives :
[ 0. 0. 0. 0. 0. 0.607 0.959 0.98 0.328 -0.287
-0.61 -0.314 -0.18 -0.8 -0.782 -0.847 -0.811 -0.825 -0.869 -0.283
0.566 0.863 0.643 0.454]
Any solution without the loop please?
CodePudding user response:
Ask and you shall receive. Here is a solution that uses recursion:
import numpy as np
data = np.array([10,5,8,9,15,22,26,11,15,16,18,7,4,8,-2,-3,-4,-6,-2,0,10,0,5,8])
length = 5
def rolling_correlation_recurse(data, i, length) :
assert i length < data.size
left = np.array([np.corrcoef(data[i:i length], np.arange(length))[0, 1]])
if i length 1 == data.size :
return left
right = rolling_correlation_recurse(data, i 1, length)
return np.concatenate((left, right))
def rolling_correlation(data, length) :
return np.concatenate((
np.zeros([length,]),
rolling_correlation_recurse(data, 0, length)
))
print(rolling_correlation(data, length))
Edit: here is a numpy solution too:
n = len(data)
print(np.corrcoef(np.lib.stride_tricks.sliding_window_view(data, length), np.tile(np.arange(length), (n-length 1, 1)))[:n-length 1, -1])
CodePudding user response:
Use a numpy.lib.stride_tricks.sliding_window_view (available in numpy v1.20.0 )
swindow = np.lib.stride_tricks.sliding_window_view(data, (length,))
which gives a view on the data array that looks like so:
array([[10, 5, 8, 9, 15],
[ 5, 8, 9, 15, 22],
[ 8, 9, 15, 22, 26],
[ 9, 15, 22, 26, 11],
[15, 22, 26, 11, 15],
[22, 26, 11, 15, 16],
[26, 11, 15, 16, 18],
[11, 15, 16, 18, 7],
[15, 16, 18, 7, 4],
[16, 18, 7, 4, 8],
[18, 7, 4, 8, -2],
[ 7, 4, 8, -2, -3],
[ 4, 8, -2, -3, -4],
[ 8, -2, -3, -4, -6],
[-2, -3, -4, -6, -2],
[-3, -4, -6, -2, 0],
[-4, -6, -2, 0, 10],
[-6, -2, 0, 10, 0],
[-2, 0, 10, 0, 5],
[ 0, 10, 0, 5, 8]])
Now, we want to apply the correlation coefficient calculation to each row of this array. Unfortunately, np.corrcoef doesn't take an axis argument, it applies the calculation to the entire matrix and doesn't provide a way to do so for each row/column.
However, the calculation for the correlation coefficient of two vectors is quite simple:
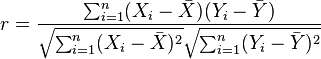
Applying that here:
def vec_corrcoef(X, y, axis=1):
Xm = np.mean(X, axis=axis, keepdims=True)
ym = np.mean(y)
n = np.sum((X - Xm) * (y - ym), axis=axis)
d = np.sqrt(np.sum((X - Xm)**2, axis=axis) * np.sum((y - ym)**2))
return n / d
Now, call this function with our array and arange:
cc = vec_corrcoef(swindow, np.arange(length))
which gives the desired result:
array([ 0.60697698, 0.95894955, 0.98 , 0.3279521 , -0.28709766,
-0.61035663, -0.31390158, -0.17995394, -0.80041656, -0.78192905,
-0.84702587, -0.81091772, -0.82464375, -0.86892667, -0.28347335,
0.56568542, 0.86304424, 0.64326752, 0.45374261, 0.38135638])
To get your x, just set the appropriate indices of a zeros array of the correct size.
Note: I think your x should contain nonzero values starting at the 4 index (because that's where the sliding window is full) instead of starting at index 5.
x = np.zeros(data.shape)
x[-len(cc):] = cc
If you are sure that your values should start at the index 5, then you can do:
x = np.zeros(data.shape)
x[length:] = cc[:-1] # Ignore the last value in cc
Comparing the runtimes of your original approach with those suggested in the answers here:
f_OP_loopyis your approach, which implements a sliding window using a loopf_PH_numpyis my approach, which uses thesliding_window_viewand the vectorized function for row-wise calculation of the vector correlation coefficientf_RA_numpyis Rontogiannis's approach, which tiles thearange, calculates the correlation coefficient for the entire matrices, and only selects the firstlen(data) - lengthrows of the last columnf_RA_recuris Rontogiannis's recursive approach, but I didn't time this because it misses out on the last correlation coefficient.
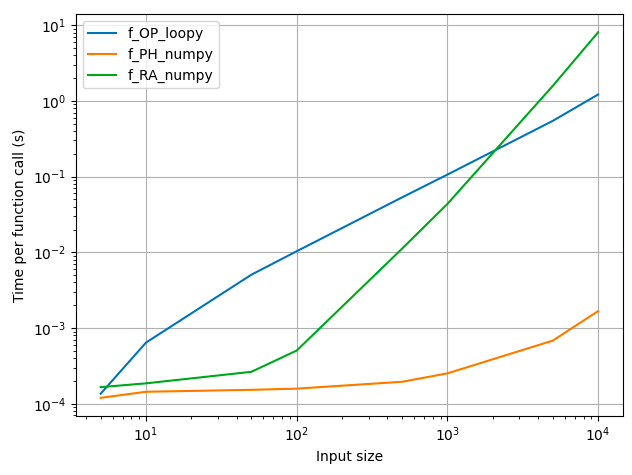
- Unsurprisingly, the numpy-only solution is faster than the loopy approach.
- My numpy solution, which computes the row-wise correlation coefficient, is faster than that shown by Rontogiannis below, because the extra work involved in tiling the vector input and calculating the correlation of the entire matrix, only to discard the unwanted elements, is avoided by my approach.
- As the input data size increases, this "extra work" in Rontogiannis's approach increases so much that its runtime is worse even than the loopy approach! I am unsure if this extra time is in the
np.corrcoefcalculation or in thenp.tileoperation.
Note: This plot was obtained on my 2.2GHz i7 Macbook Air with 8GB RAM, Python 3.10.7 and numpy 1.23.3. Similar results were obtained on Google Colab
If you're interested in the timing code, here it is:
import timeit
import numpy as np
from matplotlib import pyplot as plt
def time_funcs(funcs, sizes, arg_gen, N=20):
times = np.zeros((len(sizes), len(funcs)))
gdict = globals().copy()
for i, s in enumerate(sizes):
args = arg_gen(s)
print(args)
for j, f in enumerate(funcs):
gdict.update(locals())
try:
times[i, j] = timeit.timeit("f(*args)", globals=gdict, number=N) / N
print(f"{i}/{len(sizes)}, {j}/{len(funcs)}, {times[i, j]}")
except ValueError:
print(f"ERROR in {f}, with args=", *args)
return times
def plot_times(times, funcs):
fig, ax = plt.subplots()
for j, f in enumerate(funcs):
ax.plot(sizes, times[:, j], label=f.__name__)
ax.set_xlabel("Array size")
ax.set_ylabel("Time per function call (s)")
ax.set_xscale("log")
ax.set_yscale("log")
ax.legend()
ax.grid()
fig.tight_layout()
return fig, ax
#%%
def arg_gen(n):
return [np.random.randint(-100, 100, (n,)), 5]
#%%
def f_OP_loopy(data, length):
x = np.zeros_like(data).astype('float32')
for i in range(length-1, data.shape[0]):
x[i] = np.corrcoef(data[i - length 1:i 1], np.arange(length))[0, 1]
return x
def f_PH_numpy(data, length):
swindow = np.lib.stride_tricks.sliding_window_view(data, (length,))
cc = vec_corrcoef(swindow, np.arange(length))
x = np.zeros(data.shape)
x[-len(cc):] = cc
return x
def f_RA_recur(data, length):
return np.concatenate((
np.zeros([length,]),
rolling_correlation_recurse(data, 0, length)
))
def f_RA_numpy(data, length):
n = len(data)
cc = np.corrcoef(np.lib.stride_tricks.sliding_window_view(data, length), np.tile(np.arange(length), (n-length 1, 1)))[:n-length 1, -1]
x = np.zeros(data.shape)
x[-len(cc):] = cc
return x
#%%
def rolling_correlation_recurse(data, i, length) :
assert i length < data.size
left = np.array([np.corrcoef(data[i:i length], np.arange(length))[0, 1]])
if i length 1 == data.size :
return left
right = rolling_correlation_recurse(data, i 1, length)
return np.concatenate((left, right))
def vec_corrcoef(X, y, axis=1):
Xm = np.mean(X, axis=axis, keepdims=True)
ym = np.mean(y)
n = np.sum((X - Xm) * (y - ym), axis=axis)
d = np.sqrt(np.sum((X - Xm)**2, axis=axis) * np.sum((y - ym)**2))
return n / d
#%%
if __name__ == "__main__":
#%% Set up sim
sizes = [5, 10, 50, 100, 500, 1000, 5000, 10_000] #, 50_000, 100_000]
funcs = [f_OP_loopy, #f_RA_recur,
f_PH_numpy, f_RA_numpy]
#%% Run timing
time_fcalls = np.zeros((len(sizes), len(funcs))) * np.nan
time_fcalls = time_funcs(funcs, sizes, arg_gen)
fig, ax = plot_times(time_fcalls, funcs)
ax.set_xlabel(f"Input size")
plt.show()
input("Enter x to exit")
