I've uploaded my data onto github here: 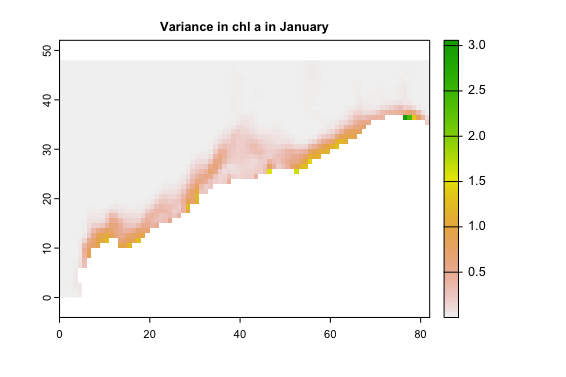
As you can see the latitude and longitude are the wrong way round. How do I fix this?
CodePudding user response:
You can open the file like this:
library(terra)
#terra 1.6.47
f <- "cmems_mod_glo_bgc_my_0.25_P1M-m_1673881119174.nc"
r <- rast(f)
Inspect
r
#class : SpatRaster
#dimensions : 82, 48, 936 (nrow, ncol, nlyr)
#resolution : 0.25, 0.25 (x, y)
#extent : 8.875, 20.875, -35.875, -15.375 (xmin, xmax, ymin, ymax)
#coord. ref. : lon/lat WGS 84
#source : cmems_mod_glo_bgc_my_0.25_P1M-m_1673881119174.nc
#varname : chl (Total Chlorophyll)
#names : chl_d~001_1, chl_d~855_1, chl_d~819_1, chl_d~001_2, chl_d~855_2, chl_d~819_2, ...
#unit : mg m-3, mg m-3, mg m-3, mg m-3, mg m-3, mg m-3, ...
#time : 1994-01-16 12:00:00 to 2019-12-16 12:00:00 UTC
names(r)[1:3]
#[1] "chl_depth=0.50576001_1" "chl_depth=1.555855_1" "chl_depth=2.6676819_1"
You can get the layer numbers for the depth you want like this
i <- grep("chl_depth=0.5", names(r))
#or like this
i <- seq(1, nlyr(r), 3)
And subset SpatRaster r
r1 <- r[[i]]
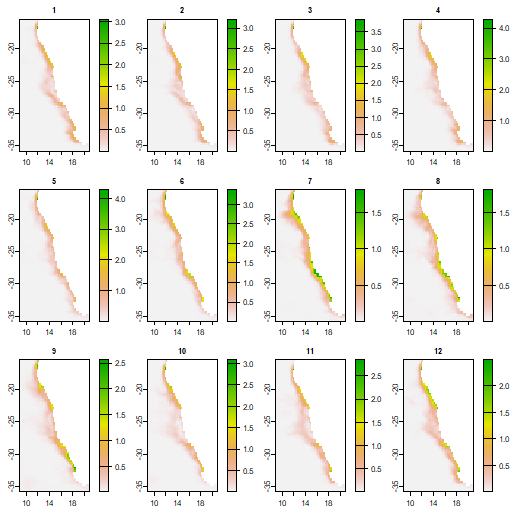
To get the variance within each month, you can now do
vm <- tapp(r1, "month", var)
vm
#class : SpatRaster
#dimensions : 82, 48, 12 (nrow, ncol, nlyr)
#resolution : 0.25, 0.25 (x, y)
#extent : 8.875, 20.875, -35.875, -15.375 (xmin, xmax, ymin, ymax)
#coord. ref. : lon/lat WGS 84
#source(s) : memory
#names : m_1, m_2, m_3, m_4, m_5, m_6, ...
#min values : 0.0001622432, 2.360285e-05, 1.136451e-05, 4.821852e-05, 0.0002333084, 0.0009578676, ...
#max values : 3.0582257258, 3.222419e 00, 3.880621e 00, 4.295268e 00, 4.2899640566, 3.3797369950, ...
#time (mnts) : Jan to Dec
plot(vm)
Doing things "by hand" like you do is more error-prone, but you could fix your result by doing
vcj <- flip(t(variance_chl_jan), "v")