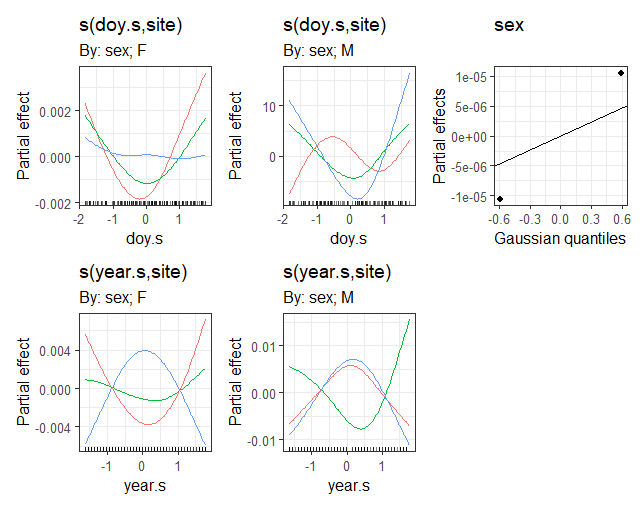
I have a dataset that I am modeling with a gam. Because there are two continuous varaibles in the gam, I have centred and scaled these variables before adding them to the model. Therefore, when I use the built-in features in gratia to show the results, the x values are not the same as the original scale. I'd like to plot the results using the scale of the original data.
An example:
library(tidyverse)
library(mgcv)
library(gratia)
set.seed(42)
df <- data.frame(
doy = sample.int(90, 300, replace = TRUE),
year = sample(c(1980:2020), size = 300, replace = TRUE),
site = c(rep("A", 150), rep("B", 80), rep("C", 70)),
sex = sample(c("F", "M"), size = 300, replace = TRUE),
mass = rnorm(300, mean = 500, sd = 50)) %>%
mutate(doy.s = scale(doy, center = TRUE, scale = TRUE),
year.s = scale(year, center = TRUE, scale = TRUE),
across(c(sex, site), as.factor))
m1 <- gam(mass ~
s(year.s, site, bs = "fs", by = sex, k = 5)
s(doy.s, site, bs = "fs", by = sex, k = 5)
s(sex, bs = "re"),
data = df, method = "REML", family = gaussian)
draw(m1)
How do I re-plot the last two panels in this figure to show the relationship between year and mass with ggplot?
CodePudding user response:
You can't do this with gratia::draw automatically (unless I'm mistaken).* But you can use gratia::smooth_estimates to get a dataframe which you can then do whatever you like with.
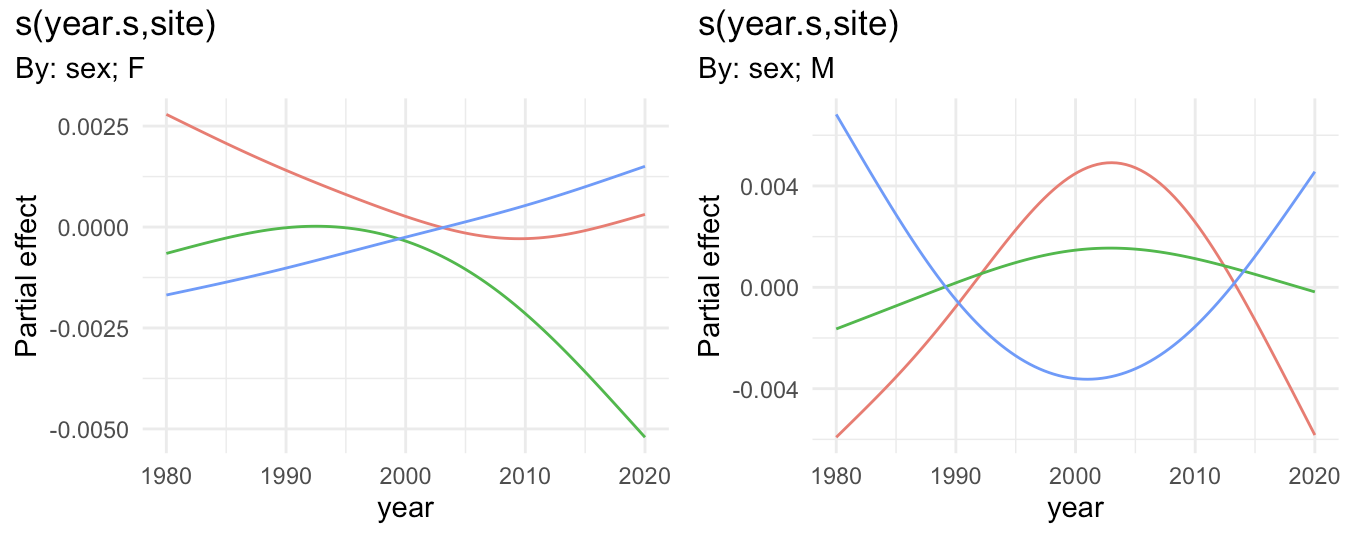
To answer your specific question: to re-plot the last two panels of the plot you provided, but with year unscaled, you can do the following
# Get a tibble of smooth estimates from the model
sm <- gratia::smooth_estimates(m1)
# Add a new column for the unscaled year
sm <- sm %>% mutate(year = mean(df$year) (year.s * sd(df$year)))
# Plot the smooth s(year.s,site) for sex=F with year unscaled
pF <- sm %>% filter(smooth == "s(year.s,site):sexF" ) %>%
ggplot(aes(x = year, y = est, color=site))
geom_line()
theme(legend.position = "none")
labs(y = "Partial effect", title = "s(year.s,site)", subtitle = "By: sex; F")
# Plot the smooth s(year.s,site) for sex=M with year unscaled
pM <- sm %>% filter(smooth == "s(year.s,site):sexM" ) %>%
ggplot(aes(x = year, y = est, color=site))
geom_line()
theme(legend.position = "none")
labs(y = "Partial effect", title = "s(year.s,site)", subtitle = "By: sex; M")
library(patchwork) # use `patchwork` just for easy side-by-side plots
pF pM
to get:
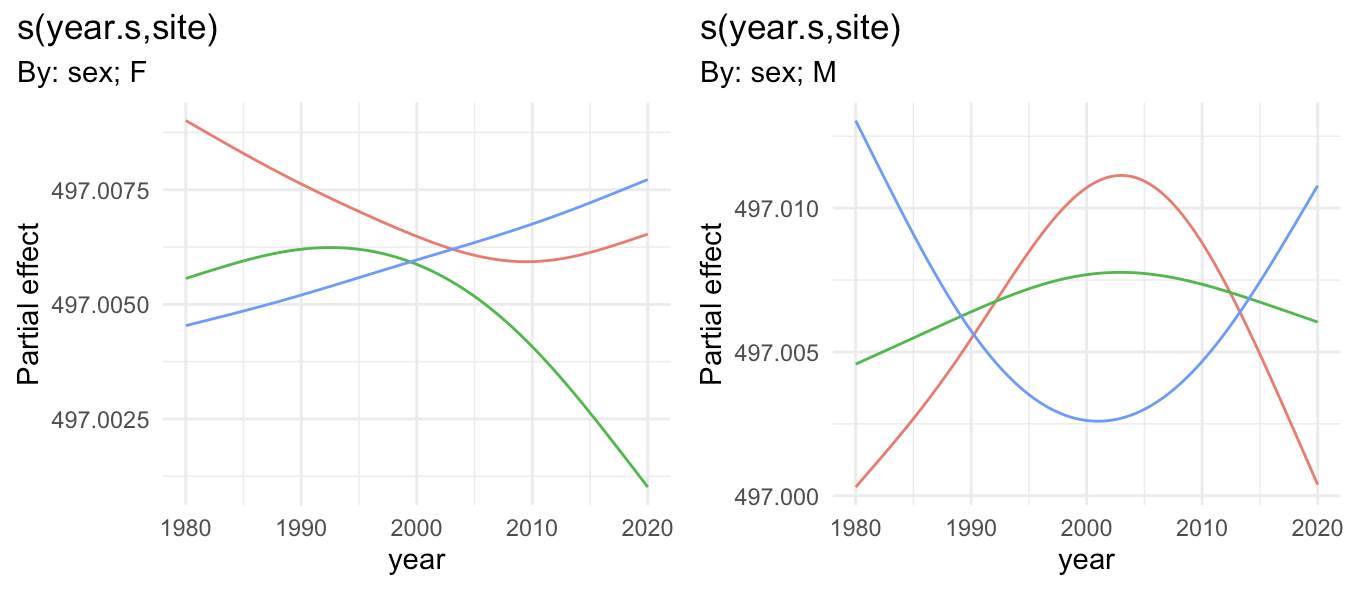
EDIT: If you also want to shift result on the y-axis as @GavinSimpson (who is the author and maintainer of gratia) mentioned, you can do this with add_constant, adding this code before plotting above:
sm <- sm %>%
add_constant(coef(m1)["(Intercept)"]) %>%
transform_fun(inv_link(m1))
[You should also in general untransform the smooth by the inverse of the model's link function. In your case this is just the identity, so it is not necessary, but in general it would be. That's what the second step above is doing.]
In your example, this results in:
*As mentioned in the custom-plotting vignette for gratia, the goal of draw not to be fully customizable, but just to be useful default. See there for recommendations about custom plots.