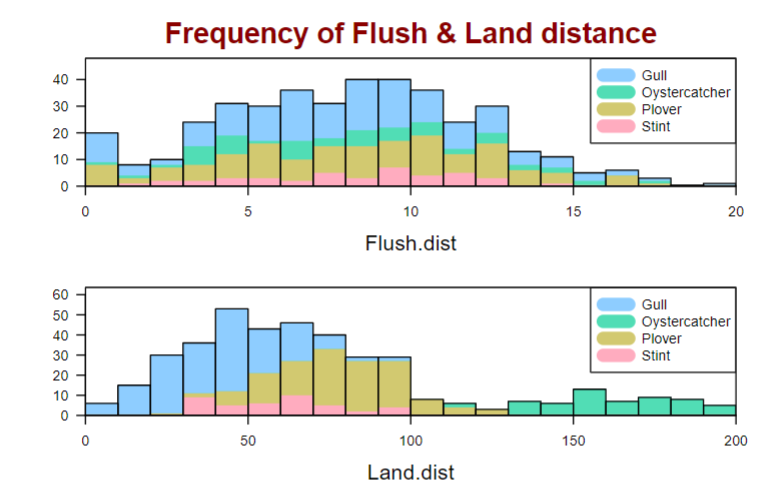
Hi I am struggling to create the below stratified histograms using a function base on a csv dataset called 'Beach-Birds.csv':
This is what I got so far:
strathist = function(x , y, breaks = 20, xlab = NULL ,ylab = NULL, main = "" , cex = 1) {
cols = hcl(h = seq(0, 360, by = 120))
h = hist(x,
breaks = 20,
plot = F)
tb = table(y, cut(x, h$breaks))
ylim <- c(-.05 * max(colSums(tb)),
1.25 * max(colSums(tb)))
barplot(
tb,
col = cols,
ylim = ylim,
ylab = ylab,
xlab = xlab,
main = main,
axisnames = FALSE,
border = NA,
space = 0
)
barplot(
colSums(tb),
ylim = ylim,
col = NA,
ylab = ylab,
xlab = xlab,
main = main,
axisnames = FALSE,
add = T,
space = 0
)
box()
legend("topright", c(rownames(tb)), fill = cols,border = NA,)
}
data=read.csv('Beach-Birds.csv')
with(data, strathist(flush.dist, Species, Sex, main = 'Frequency of Flush & Land distance'))
dput(data)
## structure(list(X = c(1L, 3L, 6L, 8L, 9L, 11L, 12L, 13L, 15L,
## 16L, 19L, 21L, 23L, 25L, 28L, 30L, 36L, 37L, 38L, 39L, 40L, 41L,
## 42L, 43L, 44L, 46L, 47L, 48L, 50L, 52L, 56L, 57L, 58L, 59L, 60L,
## 63L, 65L, 66L, 68L, 69L, 71L, 72L, 73L, 77L, 83L, 84L, 85L, 89L,
## 91L, 92L, 93L, 97L, 99L, 100L, 101L, 103L, 106L, 108L, 109L,
## 110L, 111L, 112L, 114L, 115L, 116L, 117L, 118L, 119L, 121L, 123L,
## 124L, 125L, 126L, 127L, 128L, 130L, 131L, 132L, 135L, 136L, 137L,
## 138L, 139L, 140L, 141L, 142L, 143L, 145L, 147L, 148L, 149L, 150L,
## 151L, 152L, 153L, 154L, 156L, 157L, 158L, 159L, 161L, 162L, 163L,
## 167L, 169L, 171L, 172L, 174L, 175L, 176L, 177L, 178L, 179L, 180L,
## 183L, 186L, 187L, 188L, 189L, 190L, 191L, 192L, 193L, 194L, 195L,
## 196L, 197L, 198L, 199L, 200L, 201L, 202L, 203L, 204L, 205L, 206L,
## 207L, 208L, 209L, 210L, 211L, 212L, 213L, 214L, 217L, 220L, 223L,
## 224L, 225L, 226L, 227L, 228L, 229L, 232L, 236L, 237L, 239L, 240L,
## 241L, 244L, 245L, 247L, 250L, 251L, 252L, 253L, 254L, 257L, 259L,
## 260L, 261L, 263L, 264L, 265L, 266L, 267L, 268L, 269L, 270L, 271L,
## 273L, 275L, 276L, 277L, 278L, 279L, 280L, 282L, 283L, 287L, 290L,
## 291L, 292L, 293L, 294L, 296L, 297L, 298L, 300L, 301L, 304L, 305L,
## 306L, 308L, 309L, 310L, 311L, 314L, 315L, 316L, 318L, 321L, 322L,
## 324L, 325L, 327L, 328L, 329L, 330L, 331L, 332L, 334L, 336L, 338L,
## 341L, 342L, 343L, 346L, 347L, 348L, 350L, 351L, 353L, 355L, 356L,
## 360L, 361L, 362L, 364L, 365L, 366L, 369L, 370L, 373L, 375L, 377L,
## 378L, 379L, 380L, 381L, 382L, 384L, 385L, 386L, 387L, 388L, 390L,
## 391L, 392L, 394L, 397L, 399L, 400L, 402L, 403L, 405L, 406L, 407L,
## 409L, 412L, 413L, 414L, 417L, 418L, 419L, 420L, 421L, 422L, 423L,
## 425L, 428L, 429L, 430L, 431L, 432L, 433L, 434L, 435L, 437L, 438L,
## 439L, 440L, 442L, 444L, 448L, 450L, 451L, 456L, 458L, 461L, 462L,
## 463L, 464L, 465L, 466L, 469L, 470L, 471L, 472L, 475L, 476L, 477L,
## 478L, 479L, 480L, 483L, 484L, 485L, 486L, 487L, 488L, 489L, 490L,
## 492L, 493L, 497L, 502L, 504L, 505L, 508L, 509L, 510L, 511L, 512L,
## 517L, 519L, 520L, 523L, 524L, 525L, 527L, 528L, 529L, 530L, 531L,
## 533L, 536L, 537L, 538L, 539L, 541L, 543L, 545L, 548L, 551L, 552L,
## 553L, 555L, 557L, 558L, 562L, 564L, 566L, 568L, 569L, 573L, 574L,
## 576L, 578L, 579L, 580L, 581L, 586L, 587L, 588L, 591L, 593L, 594L,
## 595L, 596L, 598L, 599L, 601L, 602L, 606L, 607L, 608L, 611L, 612L,
## 614L, 616L, 618L, 620L, 621L, 623L, 624L, 625L, 627L, 628L),
## Site = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
## 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L,
## 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
## 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
## 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
## 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
## 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
## 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L,
## 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
## 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
## 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
## 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L,
## 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 1L, 1L,
## 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
## 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
## 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 1L, 1L, 1L, 1L, 1L, 1L,
## 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
## 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
## 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
## 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
## 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
## 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
## 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L,
## 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
## 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L,
## 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
## 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L), Species = c("Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Oystercatcher", "Oystercatcher", "Oystercatcher", "Oystercatcher",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Plover", "Plover", "Plover", "Plover", "Plover",
## "Plover", "Stint", "Stint", "Stint", "Stint", "Stint", "Stint",
## "Stint", "Stint", "Stint", "Stint", "Stint", "Stint", "Stint",
## "Stint", "Stint", "Stint", "Stint", "Stint", "Stint", "Stint",
## "Stint", "Stint", "Stint", "Stint", "Stint", "Stint", "Stint",
## "Stint", "Stint", "Stint", "Stint", "Stint", "Stint", "Stint",
## "Stint", "Stint", "Stint", "Stint", "Stint", "Stint", "Stint",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull",
## "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull", "Gull"
## ), Sex = c("Female", "Male", "Female", "Female", "Female",
## "Male", "Male", "Male", "Male", "Female", "Male", "Male",
## "Female", "Female", "Female", "Male", "Female", "Female",
## "Male", "Female", "Male", "Male", "Female", "Male", "Female",
## "Male", "Male", "Female", "Male", "Male", "Male", "Female",
## "Male", "Male", "Female", "Female", "Male", "Female", "Female",
## "Female", "Male", "Female", "Female", "Female", "Female",
## "Male", "Male", "Male", "Female", "Male", "Male", "Male",
## "Female", "Male", "Female", "Female", "Male", "Male", "Male",
## "Male", "Female", "Female", "Female", "Male", "Female", "Female",
## "Male", "Female", "Male", "Female", "Male", "Female", "Male",
## "Female", "Female", "Female", "Female", "Male", "Male", "Male",
## "Female", "Female", "Female", "Male", "Female", "Male", "Female",
## "Female", "Female", "Male", "Male", "Female", "Male", "Male",
## "Female", "Male", "Female", "Male", "Female", "Female", "Female",
## "Female", "Female", "Male", "Male", "Male", "Male", "Female",
## "Female", "Male", "Female", "Female", "Female", "Male", "Male",
## "Male", "Male", "Female", "Male", "Male", "Male", "Male",
## "Male", "Female", "Male", "Female", "Female", "Male", "Female",
## "Female", "Female", "Female", "Female", "Female", "Male",
## "Male", "Female", "Male", "Male", "Male", "Male", "Female",
## "Female", "Male", "Female", "Female", "Female", "Male", "Male",
## "Female", "Female", "Male", "Male", "Male", "Male", "Male",
## "Male", "Male", "Female", "Female", "Female", "Female", "Male",
## "Female", "Male", "Female", "Female", "Male", "Female", "Female",
## "Female", "Male", "Female", "Female", "Male", "Male", "Male",
## "Female", "Male", "Female", "Male", "Female", "Male", "Female",
## "Male", "Female", "Female", "Female", "Male", "Female", "Female",
## "Female", "Male", "Female", "Male", "Male", "Male", "Male",
## "Female", "Female", "Male", "Male", "Female", "Female", "Female",
## "Male", "Female", "Female", "Female", "Male", "Female", "Female",
## "Female", "Female", "Female", "Male", "Male", "Male", "Female",
## "Female", "Female", "Female", "Female", "Female", "Female",
## "Male", "Female", "Male", "Female", "Male", "Male", "Male",
## "Male", "Female", "Male", "Female", "Male", "Male", "Male",
## "Female", "Male", "Male", "Female", "Male", "Female", "Male",
## "Male", "Female", "Female", "Female", "Male", "Female", "Male",
## "Male", "Female", "Female", "Male", "Female", "Male", "Female",
## "Female", "Male", "Female", "Male", "Male", "Female", "Female",
## "Male", "Female", "Male", "Female", "Male", "Male", "Male",
## "Male", "Male", "Female", "Male", "Male", "Male", "Male",
## "Female", "Male", "Male", "Male", "Male", "Female", "Male",
## "Female", "Female", "Male", "Female", "Male", "Male", "Female",
## "Male", "Female", "Male", "Male", "Male", "Male", "Male",
## "Male", "Female", "Female", "Female", "Male", "Male", "Male",
## "Female", "Male", "Male", "Male", "Male", "Female", "Female",
## "Female", "Female", "Female", "Female", "Female", "Male",
## "Male", "Male", "Female", "Female", "Male", "Female", "Male",
## "Female", "Male", "Female", "Male", "Female", "Female", "Male",
## "Male", "Male", "Female", "Female", "Female", "Female", "Male",
## "Female", "Female", "Female", "Male", "Male", "Male", "Male",
## "Male", "Male", "Male", "Male", "Female", "Female", "Female",
## "Female", "Male", "Female", "Female", "Male", "Female", "Male",
## "Male", "Male", "Male", "Male", "Female", "Male", "Female",
## "Male", "Female", "Female", "Male", "Male", "Male", "Female",
## "Male", "Male", "Male", "Male", "Male", "Female", "Male",
## "Male", "Female", "Male", "Male", "Male", "Male", "Female",
## "Male", "Female", "Female", "Male", "Male", "Female", "Female"
## ), flush.dist = c(12.8, 4.4, 9.5, 12.2, 10.1, 6.6, 15.4,
## 11.3, 13.8, 12.9, 9.9, 3.1, 8.2, 1.3, 8.2, 7.8, 15.1, 8.1,
## 5.8, 10.3, 9.4, 8.3, 17.7, 3.9, 6.4, 8.6, 4.5, 4.6, 10.1,
## 4.5, 3.5, 7.3, 4.8, 10.3, 10.2, 8.9, 9.1, 3.4, 7.8, 4.7,
## 13.2, 6.3, 3.2, 7, 3.9, 6.8, 9.8, 6.9, 2.4, 0.1, 14.8, 3.5,
## 4.6, 12, 12.2, 14.3, 6.7, 6.4, 6.3, 5.5, 17.8, 10.4, 14.2,
## 4.7, 5.7, 9.8, 13.7, 12.2, 13.9, 7.7, 12.2, 13.9, 13, 13,
## 10.9, 7.6, 7.7, 5.7, 7.8, 2.6, 4.6, 13.8, 7.4, 4.6, 10.6,
## 13.1, 2.7, 12.8, 5.3, 8.4, 8.2, 12.2, 0.1, 9.4, 4, 11, 4.1,
## 2.2, 6.8, 0.2, 5.8, 10.9, 4.6, 5.7, 7.4, 12.5, 6.8, 5.8,
## 6.2, 0.9, 5.5, 3.3, 5.4, 2, 10.9, 14.2, 5.9, 7.3, 10.3, 16.6,
## 12, 12.9, 3, 9.9, 3, 10.7, 4.4, 10.5, 10.6, 0.1, 0.9, 12.7,
## 10.7, 12.2, 16.3, 9.7, 9.8, 16.1, 6.7, 14.8, 7.5, 10.8, 14.7,
## 13.5, 5.9, 5.3, 8.4, 11, 4.2, 12.6, 8.8, 11.6, 11.1, 11,
## 13, 8.2, 11.1, 4, 8.8, 0.8, 9, 11.5, 9.3, 8.4, 7.3, 8.2,
## 8, 8.5, 3.1, 12.8, 11.9, 3.3, 1.3, 1, 6.9, 9.4, 0.1, 10,
## 6.1, 8.3, 3.2, 10.7, 5.9, 11.4, 8.3, 16.4, 4.8, 4.1, 9.9,
## 9.6, 3.2, 7.1, 11.7, 8, 11.6, 12.2, 3.6, 9.3, 4.4, 10.4,
## 7.3, 12.6, 5.5, 11.7, 9.1, 7.5, 9.5, 12.4, 10.1, 9.8, 4.7,
## 11.6, 7, 9, 2.7, 5.3, 6.7, 8.7, 5.7, 4.9, 9.4, 9.6, 10.2,
## 3, 10.2, 8.2, 14.8, 1.4, 7.2, 11.3, 9.2, 6, 7, 10.9, 4, 10.9,
## 6.9, 4.9, 6.9, 3.3, 12.6, 1.4, 8.6, 8.6, 3.4, 13.3, 5.1,
## 8.9, 4.3, 5.5, 9.4, 10, 0.1, 15.8, 6.8, 10, 4.7, 13.4, 5.3,
## 3.3, 13.2, 6.8, 13, 4.5, 14.6, 9.4, 10.1, 7.3, 9.5, 14.1,
## 11.1, 20, 5.9, 6.4, 0.1, 7.5, 4.5, 3.1, 14.6, 8.7, 13.1,
## 0.1, 8.5, 10.5, 9.1, 6.7, 3.4, 10.2, 11.9, 12.1, 7.9, 5.9,
## 9, 14.7, 12.7, 5.6, 6.5, 0.1, 12.7, 12.1, 9.3, 6.3, 7.6,
## 7.6, 8.8, 8.2, 9, 7.8, 8.4, 6.8, 6.6, 3.5, 8.6, 0.5, 4.4,
## 6.7, 3.2, 15.2, 5.5, 0.8, 8.3, 17.2, 9.2, 4.6, 8.8, 0.2,
## 8.5, 0.8, 5.1, 8.8, 9.4, 8.4, 10.6, 9.9, 7.6, 5.2, 6.2, 1.6,
## 6.7, 7.6, 7.9, 17, 12.1, 13.9, 6.7, 11.8, 6.9, 5.1, 9.4,
## 7.3, 11.2, 12.7, 4.4, 4.9, 12.5, 4.5, 7.3, 11.6, 9.1, 11.1,
## 10.1, 6.3, 13, 9.7, 9.2, 11.2, 2.6, 9.3, 8.6, 0, 11.7, 10.1,
## 10.2, 9.5, 0.1, 9, 5.7, 15.4, 7.2, 0, 16.7, 11, 10.4, 6.7,
## 6, 4.9, 8.9, 1.1, 11.3, 1.1, 3.5, 2.1, 9.6, 11.8, 4.8, 6.2,
## 7.6, 9.4, 10.1), land.dist = c(150.9, 114.1, 153.7, 137.7,
## 143.3, 142.8, 134.1, 159.5, 150.4, 161.9, 136.3, 142.2, 157.3,
## 153.4, 154.7, 142, 135.8, 163.1, 138.2, 116.7, 152, 136,
## 190.7, 179.1, 157.6, 155.5, 180, 177.3, 162, 162.6, 142.3,
## 182.8, 182.7, 168.7, 156.6, 160.3, 136.4, 175.8, 169, 170.4,
## 150.5, 159.3, 173.7, 147.7, 197.2, 174.5, 184.6, 188.4, 185.7,
## 174.3, 199.9, 186.4, 198.9, 193.5, 187, 187.2, 178.5, 58.6,
## 62.5, 88.3, 79.3, 48.8, 70.3, 66.5, 40.5, 55, 43.9, 50.6,
## 66.9, 27.8, 89.3, 62.4, 74.8, 56.5, 62.1, 74.9, 56.9, 52.3,
## 69.8, 68.7, 96.7, 58, 65.5, 53.6, 53.6, 77.7, 97.5, 80.9,
## 54.7, 36.4, 77.9, 81.3, 96.8, 79.8, 82.8, 75.8, 84.6, 73.9,
## 49, 97.7, 72.1, 94.3, 60, 52, 46.9, 62.3, 86.6, 70.1, 38.5,
## 92.1, 83.8, 58.6, 45.3, 64.4, 79, 66.2, 88.6, 83.4, 98.6,
## 72.1, 91.1, 84, 77.9, 64.7, 77.8, 93.2, 94.6, 74.7, 99.2,
## 69, 116.3, 96, 126.1, 82.6, 89.3, 102.9, 77.4, 76.3, 93.9,
## 88.5, 111.5, 90.1, 100.5, 103.2, 89, 97.9, 92.4, 75.6, 80.4,
## 90, 88.5, 55.8, 81.7, 72.6, 64.7, 43.7, 73.5, 74.1, 68.3,
## 79.6, 81.9, 64.7, 80.9, 87.3, 78.7, 58.7, 85.8, 72.7, 93.2,
## 77.2, 73.5, 60.7, 100.6, 125.2, 115.3, 83.9, 107.9, 99.1,
## 97.4, 93.4, 89.5, 113.3, 108.4, 108.6, 99.8, 99.6, 128.8,
## 93.4, 76.5, 103.4, 30.4, 62.6, 65.1, 55.6, 34.1, 44.7, 48.1,
## 37.7, 35.7, 37.6, 59.5, 67.4, 55, 65.2, 37.6, 39, 46, 41.4,
## 63.3, 66.5, 82.9, 66.7, 61.9, 90.1, 76.9, 73.3, 36.8, 61.1,
## 57, 54, 60.2, 53.6, 43, 38, 90.3, 93.7, 70.3, 72.5, 97.3,
## 81.3, 75.6, 30, 34.8, 19.4, 6.7, 15.3, 6.4, 10.8, 29.4, 5.6,
## 19.1, 9.1, 21, 24.1, 13.4, 44.3, 33.3, 26.7, 7.9, 12.5, 17.5,
## 37.5, 41.6, 20.2, 12.4, 7.6, 16.5, 54.6, 24.1, 35.2, 20.1,
## 29.4, 16.7, 25.3, 25.1, 47.6, 27, 35.2, 26.4, 43.7, 33.3,
## 46.4, 32.5, 33, 41.6, 31.4, 18.4, 32.3, 48.9, 18.8, 53.1,
## 52.1, 30, 16.9, 45.1, 37.3, 22.8, 38, 47.3, 57.5, 52.7, 15.9,
## 33.6, 21.3, 40.8, 51, 52.6, 21, 37.3, 27.9, 19.8, 69.4, 31.1,
## 25.6, 62.2, 57.4, 36.5, 54.7, 41.7, 38.3, 50.4, 56.7, 92.8,
## 37, 67.3, 67.5, 71.4, 48.2, 54.1, 43, 47, 55.7, 45, 51.1,
## 42.6, 61.1, 44.4, 72.1, 46, 43.6, 44.1, 42.4, 67.7, 64.5,
## 41.1, 43, 40.1, 48, 48.2, 52.2, 41.9, 37.1, 28.6, 21, 26.2,
## 21.6, 33, 21.2, 28.2, 45.4, 36.1, 24.2, 38.3, 26.3, 49.1,
## 42.4, 44.6, 32.3, 27.3, 35.2, 49.5, 34.7, 45.9, 41.9, 40.8,
## 20.6, 51.6, 83.1, 64.9, 71.6, 48, 50, 83.6, 49.4, 52.4, 69.7,
## 60.8, 64.6, 51.9, 63.6, 64, 71, 61, 51.5, 49.4, 68, 45.4,
## 71.1, 49.8, 64.7, 76.3, 72.1, 66.7, 69.4, 91.9, 66.1, 59.8,
## 50.5, 52.1)), class = "data.frame", row.names = c(NA, -399L
## ))
dput() shows my dataset. As cazman requested.
But my histogram looks much different:
Would love some help. Thanks. The csv file looks something like this:
CodePudding user response:
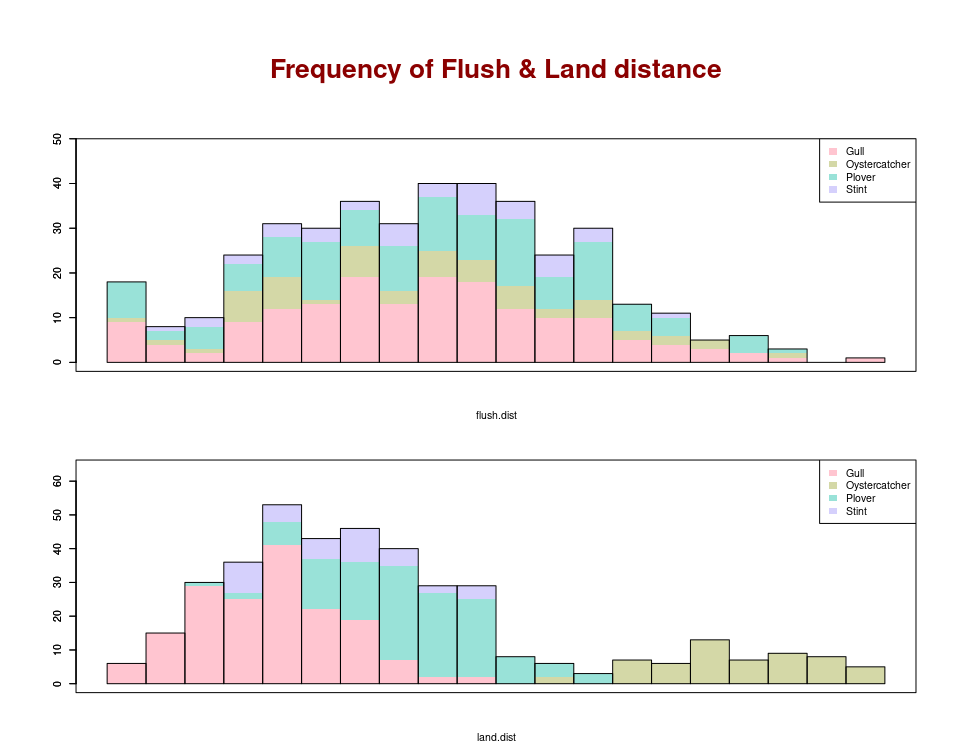
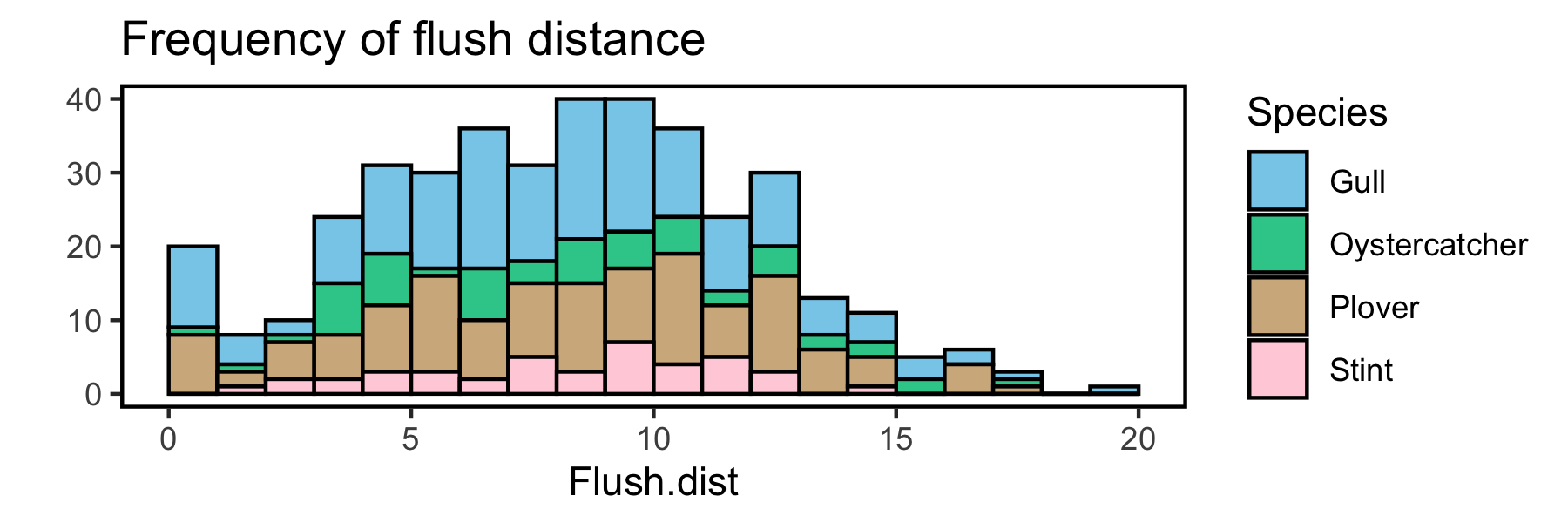
This looks pretty close to what you are after for the top graph using the package ggplot2. It's hard to know for sure without knowing how those graphs were produced.
ggplot(data = df, aes(x = flush.dist, fill = Species))
geom_histogram(bins = 20,
breaks = seq(0, 20, by = 1),
color = "black")
scale_fill_manual(values = c("#87ceeb",
"#33cc99",
"#D2B48C",
"#FFD1DC"))
xlab("Flush.dist")
ylab("")
labs(title = "Frequency of flush distance")
theme(panel.border = element_rect(fill = NA,
color = "black",
size = 1),
panel.background = element_blank())
And saved with a width of 6 inches and height of 2 inches gives:
CodePudding user response:
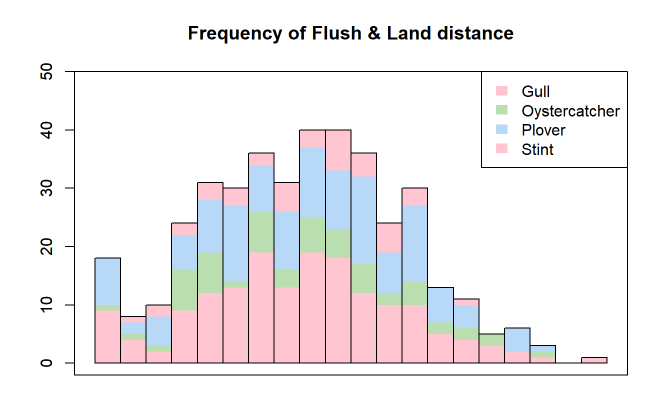
Loop over the respective column names and get their values. This should bring you pretty close.
layout(matrix(1:3, 3), heights=c(.15, .425, .425))
plot.new();text(.5, -1, "Frequency of Flush & Land distance",
cex=2.5, col='darkred', xpd=TRUE, font=2)
op <- par(mar=c(5, 6, 2, 3))
lapply(c('flush.dist', 'land.dist'), \(x) {
with(d, strathist(get(x), Species, Sex, xlab=x))
})
par(op)