This is a tricky one, and I'm making it trickier by basically asking two questions concurrently. But they're related (in practice, if not in theory).
The issue
Basically I want to use one script and one data frame to create a faceted plot with arrows:
- at unique locations on each plot and
- at consistent angles regardless of the scale.
The goal is to use the arrows to indicate dosing of a therapeutic, which might change for individuals or treatment cohorts.
Example dataset
Here's an example of the sort of data which I might need to plot with arrows:
df_3 <- tibble(
ID = rep(1:2, each = 10),
TIME = rep(1:10, times = 2),
DV = c(runif(10), runif(10) * 5)
)
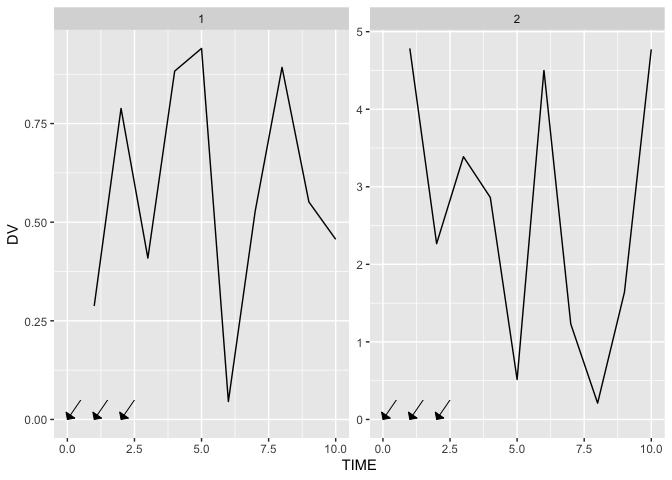
An example plot of what I DON'T want
The follow code generates an example of where I am, but not where I want to be:
ggplot(data = df_3, aes(x = TIME, y = DV))
geom_line()
facet_wrap(~ID, scales = "free_y")
annotate("segment",
x = c(0.5, 1.5, 2.5), xend = c(0, 1, 2), y = 0.05, yend = 0,
arrow = arrow(length = unit(0.20, "cm"), type = "closed"))
Note that the arrows can only be set to a single series of locations (which are tedious to add, since I need to finagle the x and xend variables to get the angle I want) and, because the y-axes are different, the angles of each set of angles is different.
For example, let's say I want arrows at times 0, 1, and 2 for Individual 1, but at times 2, 4, and 6 for Individual 2.
I'm thinking I need to add the location for the arrows into the dataset, but I'm worried that will force ggplot to plot the arrows for every individual plot, creating fuzzy/dark arrows.
I'm open to any and all suggestions or thoughts. I appreciate your time.
CodePudding user response:
The problem you're running into is that the arrows are totally defined in dataspace, which can skew the angle in visual space. One way to tackle it is to write your own geom that draws it exactly as you like, but that feels like overkill for a task that seems so simple.
This seems like a nice use case for Paul Murrell's gggrid package. One of the possibilities is to create a function that takes the raw data and transformed data (called coords), and outputs the desired graphical object.
# devtools::install_github("pmur002/gggrid")
library(ggplot2)
library(gggrid)
#> Loading required package: grid
df_3 <- data.frame(
ID = rep(1:2, each = 10),
TIME = rep(1:10, times = 2),
DV = c(runif(10), runif(10) * 5)
)
arrows <- function(data, coords) {
offset <- unit(4, "mm")
segmentsGrob(
x0 = unit(coords$x, "npc") offset,
x1 = unit(coords$x, "npc"),
y0 = unit(coords$y, "npc") offset,
y1 = unit(coords$y, "npc"),
arrow = arrow(length = unit(0.2, "cm"), type = "closed"),
gp = gpar(fill = "black")
)
}
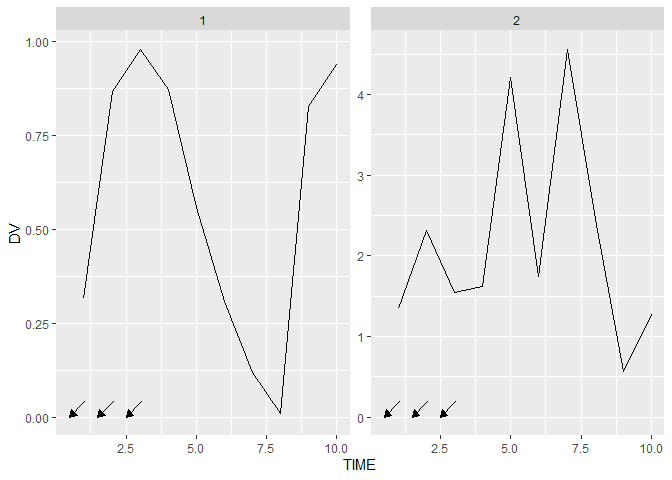
ggplot(data = df_3, aes(x = TIME, y = DV))
geom_line()
facet_wrap(~ID, scales = "free_y")
grid_panel(data = data.frame(TIME = c(0.5, 1.5, 2.5), DV = 0),
grob = arrows)
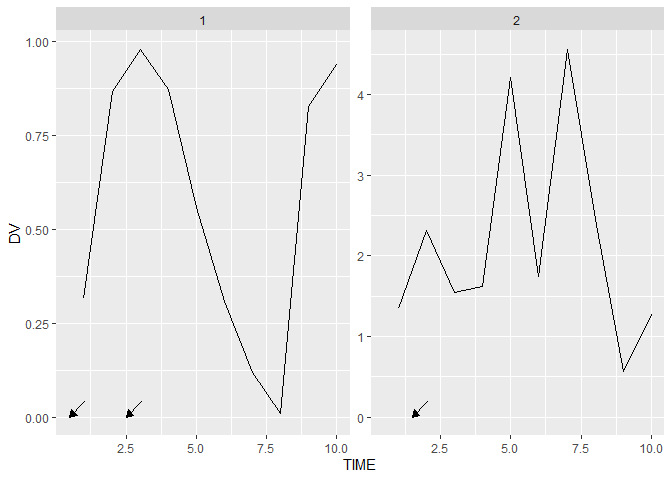
The nice thing is that this is not a static annotation such as annotation_custom() which is simply repeated along facets. In the example below we can see that the second arrow gets send to the second panel.
ggplot(data = df_3, aes(x = TIME, y = DV))
geom_line()
facet_wrap(~ID, scales = "free_y")
grid_panel(data = data.frame(TIME = c(0.5, 1.5, 2.5),
DV = 0, ID = c(1, 2, 1)), # <- facet var
grob = arrows)
Created on 2021-09-21 by the reprex package (v2.0.1)
CodePudding user response:
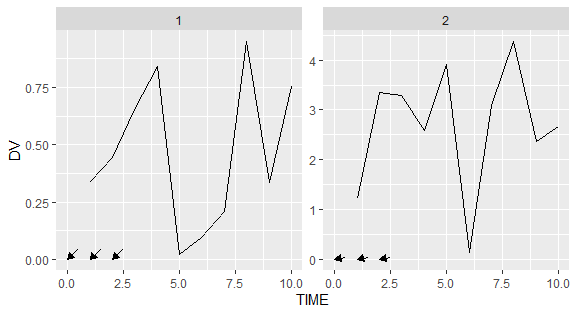
Not sure whether I fully understand what you are trying to achieve but one option to add your arrows would be to make use of annotation_custom instead of annotate.
set.seed(123)
library(tibble)
df_3 <- tibble(
ID = rep(1:2, each = 10),
TIME = rep(1:10, times = 2),
DV = c(runif(10), runif(10) * 5)
)
library(grid)
library(ggplot2)
segment <- segmentsGrob(x0 = unit(.05, "npc"), y0 = unit(.05, "npc"),
x1 = unit(0, "npc"), y1 = unit(0, "npc"),
arrow = arrow(angle = 45, length = unit(0.2, "cm"), type = "closed"), gp = gpar(fill = "black"))
ggplot(data = df_3, aes(x = TIME, y = DV))
geom_line()
scale_x_continuous(limits = c(0, NA))
scale_y_continuous(limits = c(0, NA))
facet_wrap(~ID, scales = "free_y")
lapply(0:2, function(xmin) {
annotation_custom(grob = segment, xmin = xmin, ymin = 0, xmax = max(df_3$TIME) xmin)
})