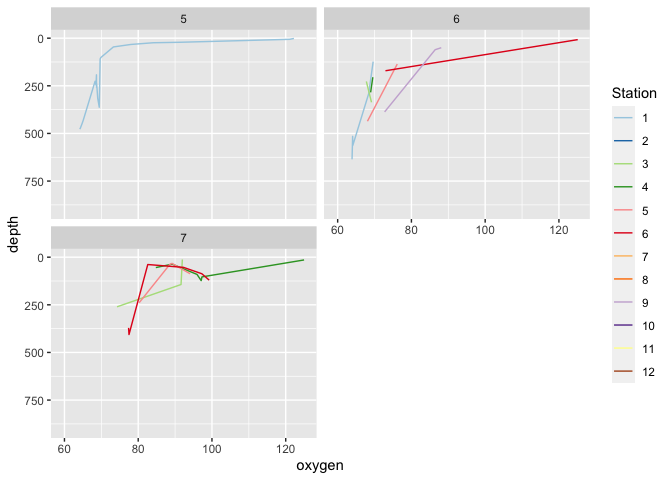
I need to represent an important number of graphs. To do that I used a the facet_wrap_paginate function, but I only have the second page printed/plotted.
Here is a sample of my data:
df <- structure(list(oxygen = c(60.86414, 62.50372, 89.76055, 61.15914,
61.49936, 61.90402, 61.60323, 61.2873, 75.12329, 60.83503, 63.03796,
61.78298, 70.16533, 62.60887, 61.49043, 64.42095, 61.78358, 62.29302,
62.88122, 60.47264, 65.92805, 65.0235, 64.32414, 65.56869, 64.65729,
66.73502, 67.01285, 67.47757, 69.62232, 66.09624, 66.18106, 64.28226,
64.85658, 67.46348, 68.93226, 65.23919, 65.67461, 64.64727, 64.90544,
68.10878, 70.00292, 59.54631, 61.43627, 78.75032, 61.94263, 82.08963,
62.75281, 61.93749, 72.86382, 62.06785, 61.04865, 95.49206, 69.79339,
66.82354, 61.53702, 85.51962, 101.53748, 61.64096, 92.14781,
60.41892, 77.55303, 75.39007, 62.15703, 82.38292, 87.27823, 69.28644,
73.72369, 62.02667, 61.34973, 61.61716, 70.61777, 61.59655, 60.31008,
61.65777, 71.82138, 61.9823, 79.38791, 60.31816, 108.2866, 61.54543,
69.87043, 64.30304, 65.05726, 68.69304, 84.0604, 121.05592, 68.67872,
69.53102, 68.31013, 64.76675, 122.22451, 91.29598, 69.43838,
68.73515, 77.99483, 68.56142, 64.14964, 73.2695, 69.67608, 69.01853,
68.5685, 63.9001, 68.93557, 71.84125, 72.73052, 69.17045, 76.28085,
68.41978, 69.53005, 76.15901, 86.43705, 72.95997, 64.0582, 70.29805,
64.10386, 88.09503, 68.07687, 69.62147, 67.77802, 125.09757,
91.96934, 97.26108, 89.16293, 80.245, 124.97964, 97.28237, 95.95074,
99.24807, 82.59685, 89.2217, 77.50349, 88.54909, 91.61343, 77.38885,
94.06567, 84.77561, 80.46941, 92.28428, 74.25416, 97.07623),
depth = c(707.5, 195, 47.5, 750, 315.5, 506.5, 660.5, 678,
146, 821.5, 197, 365, 141.5, 448, 574.5, 325.5, 645.5, 236,
446, 872.5, 315, 729.5, 786.5, 573.5, 752, 202, 455, 412,
147, 552.5, 517, 904, 339.5, 184, 308.5, 307.5, 610, 705.5,
700, 375, 228.5, 261.5, 398.5, 168.5, 360, 136.5, 504, 299.5,
187, 473.5, 249.5, 64.5, 178.5, 251.5, 369.5, 95, 66.5, 599.5,
72, 474, 178, 33.5, 109, 86.5, 26.5, 240, 51.5, 367, 295.5,
408.5, 244.5, 106, 314, 304.5, 69.5, 361.5, 91.5, 125.5,
11, 281.5, 101.5, 472.5, 433, 257, 24, 6, 192.5, 265, 226,
449, 1.5, 21.5, 362.5, 238.5, 33, 246, 477.5, 46, 109, 315,
275.5, 636, 284.5, 267, 387, 336.5, 225.5, 291, 205, 136,
60.5, 171, 515.5, 260.5, 563, 50, 436, 123, 227, 7.5, 12,
87, 32.5, 239, 14.5, 103.5, 90.5, 120.5, 38.5, 36.5, 406,
38.5, 143.5, 371, 87, 54.5, 234, 53.5, 260.5, 123), ctd_file = c("BA16007",
"BA16002", "BA16011", "BA16003", "BA16004", "BA16006", "BA16001",
"BA16002", "BA16012", "BA16003", "BA16004", "BA16004", "BA16005",
"BA16004", "BA16009", "BA16005", "BA16012", "BA16004", "BA16006",
"BA16003", "BA16013", "BA16014", "BA16017", "BA16016", "BA16018",
"BA16016", "BA16014", "BA16020", "BA16020", "BA16020", "BA16016",
"BA16018", "BA16016", "BA16018", "BA16021", "BA16015", "BA16014",
"BA16013", "BA16015", "BA16021", "BA16025", "BA16023", "BA16024",
"BA16025", "BA16024", "BA16025", "BA16025", "BA16025", "BA16024",
"BA16025", "BA16023", "BA16025", "BA16023", "BA16025", "BA16024",
"BA16022", "BA16022", "BA16025", "BA16024", "BA16024", "BA16026",
"BA16035", "BA16030", "BA16035", "BA16031", "BA16026", "BA16032",
"BA16031", "BA16027", "BA16032", "BA16035", "BA16028", "BA16031",
"BA16027", "BA16033", "BA16032", "BA16035", "BA16028", "BA16027",
"BA16032", "BA16034", "BA16034", "BA16034", "BA16034", "BA16034",
"BA16034", "BA16034", "BA16034", "BA16034", "BA16034", "BA16034",
"BA16034", "BA16034", "BA16034", "BA16034", "BA16034", "BA16034",
"BA16034", "BA16034", "BA16034", "BA16040", "BA16036", "BA16043",
"BA16047", "BA16048", "BA16041", "BA16049", "BA16036", "BA16042",
"BA16044", "BA16048", "BA16045", "BA16039", "BA16046", "BA16036",
"BA16048", "BA16044", "BA16038", "BA16041", "BA16045", "BA16052",
"BA16055", "BA16054", "BA16054", "BA16053", "BA16053", "BA16053",
"BA16055", "BA16055", "BA16053", "BA16055", "BA16054", "BA16052",
"BA16055", "BA16054", "BA16053", "BA16054", "BA16055", "BA16052",
"BA16053"), Transect = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L), .Label = c("1",
"2", "3", "4", "5", "6", "7"), class = "factor"), Station = structure(c(7L,
2L, 11L, 3L, 4L, 6L, 1L, 2L, 12L, 3L, 4L, 4L, 5L, 4L, 9L,
5L, 12L, 4L, 6L, 3L, 1L, 2L, 5L, 4L, 6L, 4L, 2L, 8L, 8L,
8L, 4L, 6L, 4L, 6L, 9L, 3L, 2L, 1L, 3L, 9L, 4L, 2L, 3L, 4L,
3L, 4L, 4L, 4L, 3L, 4L, 2L, 4L, 2L, 4L, 3L, 1L, 1L, 4L, 3L,
3L, 1L, 9L, 5L, 9L, 6L, 1L, 7L, 6L, 2L, 7L, 9L, 3L, 6L, 2L,
8L, 7L, 9L, 3L, 2L, 7L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 4L, 8L,
9L, 3L, 10L, 1L, 4L, 5L, 9L, 6L, 1L, 7L, 1L, 9L, 5L, 1L,
3L, 6L, 3L, 6L, 5L, 5L, 4L, 4L, 4L, 6L, 6L, 4L, 6L, 5L, 3L,
6L, 5L, 4L, 5L, 6L, 3L, 4L), .Label = c("1", "2", "3", "4",
"5", "6", "7", "8", "9", "10", "11", "12"), class = "factor")), row.names = c(NA,
-140L), groups = structure(list(Transect = structure(1:7, .Label = c("1",
"2", "3", "4", "5", "6", "7"), class = "factor"), .rows = structure(list(
1:20, 21:40, 41:60, 61:80, 81:100, 101:120, 121:140), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -7L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"))
And there is the code I used:
library(dplyr)
library(tidyverse)
library(ggforce)
i <- ceiling(
length(levels(df$Transect)) / 4) # set the number of pages
pdf("multi_page.pdf", width = 16 / 2.54, height = 12 / 2.54)
SdesGG <- df %>% #launch each time or does not work
group_by(Transect) %>% #mandatory or need to fortify
ggplot(aes(x = oxygen, y = depth, color = Station))
geom_line()
scale_color_brewer(palette = "Paired")
scale_y_reverse()
facet_wrap_paginate(~Transect, ncol = 2, nrow = 2, page = i) #ggforce
dev.off()
I have 180 000 observations, therefore it might be interesting to duplicate the number of row
lapply(df, rep, 1000)
CodePudding user response:
With the page argument you only specify
The page to draw (see
?facet_wrap_paginate)
That's why you get only the last or second page with page = i.
If you want all pages you have to loop over the pages:
library(ggplot2
library(ggforce)
i <- ceiling(
length(levels(df$Transect)) / 4) # set the number of pages
pdf("multi_page.pdf", width = 16 / 2.54, height = 12 / 2.54)
lapply(seq(i), function(page) {
SdesGG <- df %>% #launch each time or does not work
group_by(Transect) %>% #mandatory or need to fortify
ggplot(aes(x = oxygen, y = depth, color = Station))
geom_line()
scale_color_brewer(palette = "Paired")
scale_y_reverse()
facet_wrap_paginate(~Transect, ncol = 2, nrow = 2, page = page) #ggforce
})
dev.off()
#> [[1]]
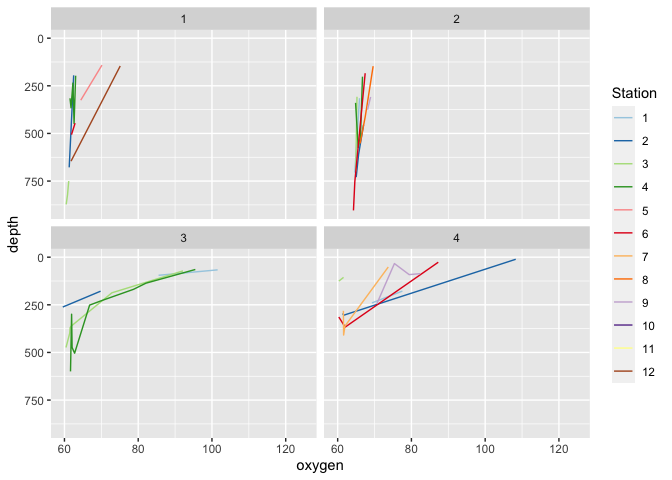
#>
#> [[2]]