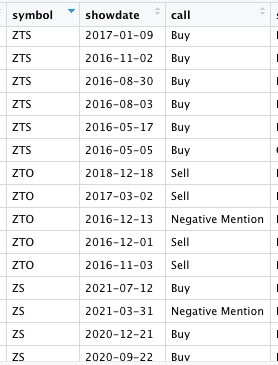
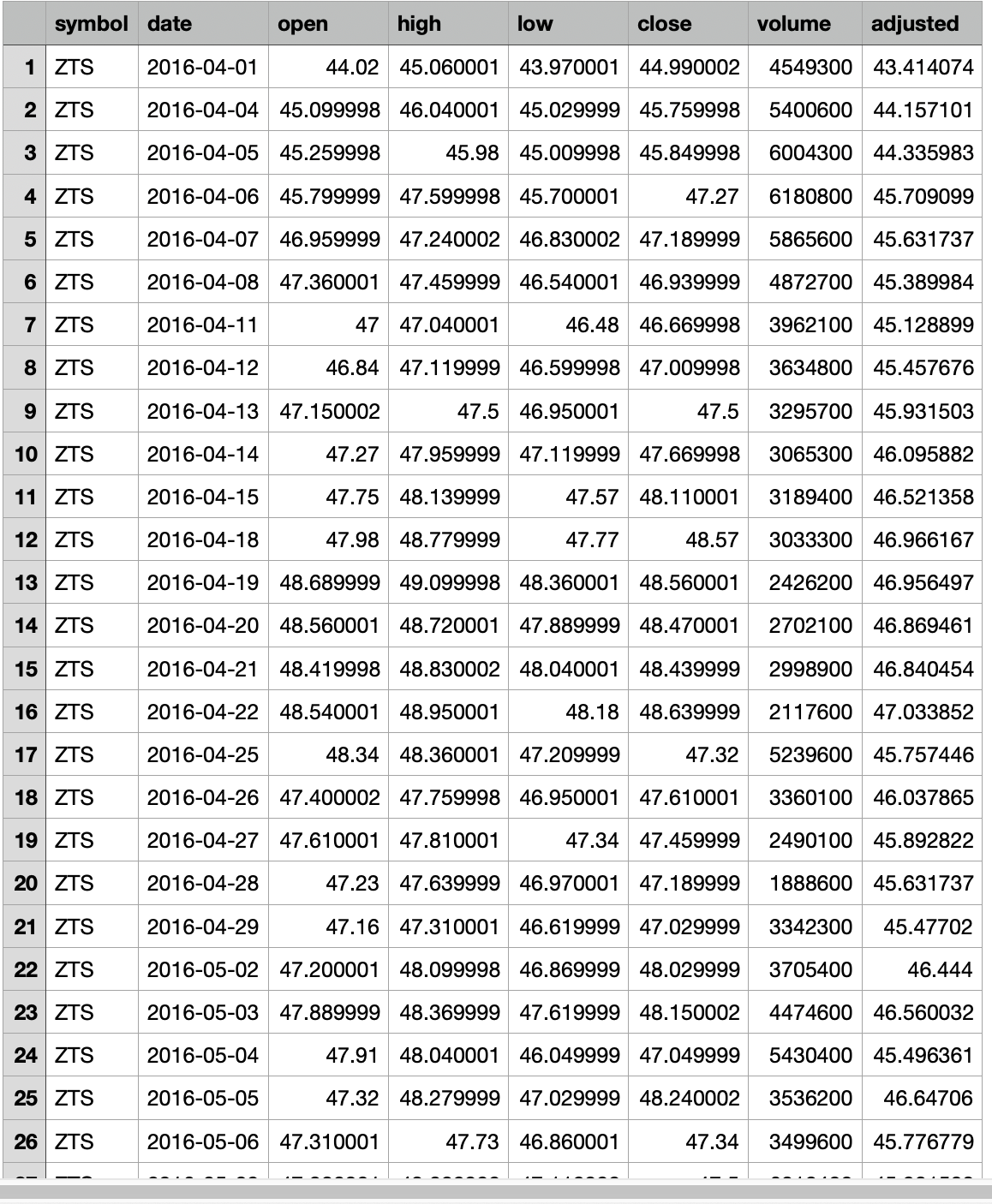
I have a table of reference stock symbol (20,000 rows)
and a folder of csv files, each CSV files is named by a stock symbol, for example ZTS.csv. Inside each csv file, is the price history of the symbol.
The end goal is to track performances of all stocks and visualisation. Because of the sheer size of the reference table and the csv files, I think the most sensible approach will be selecting need information from each CSV files and add them into the reference table.
For example, I would want to take a row from the reference table, symbol ZTS, showdate 2017-01-09,
Then read the ZTE.csv file, find the rows with date matching the showdates, add the open/high/low/close price data columns
Then loop this.
Due to size restrictions, I have uploaded sample data here on google drive: https://drive.google.com/drive/folders/1G3os67b2i2VfGHnvR6NX8qk1ECuVawGJ?usp=sharing
#read in the reference data
df <- read.csv("reference table.csv", header = TRUE)
# get csv files directory and list all files in this directory
wd <- "/Users/m/Desktop/project/price_data_csv"
files_in_wd <- list.files(wd)
#find stuff to match
# create an empty list and read in all files from wd
mylist <- list()
for(i in seq_along(files_in_wd)){
mylist[[i]] <- read.delim(file = files_in_wd[i],
sep = ',',
header = T)
}
I'm stuck on how to do the matching and creating combined table. Thank you
CodePudding user response:
I'd recommend using data.table, because, as @r2evans mentions, it does grouping well, and if your data is large, it is very fast.
Using your sample data, the could bekow should hopefully get you started (I've prefixed the data.table methods to help indicate where it's being used). You can use the provided function on an individual symbol, or try running it all at once (not sure how big your data actually is).
library(data.table)
data_dir <- "~/Downloads/Testing/"
reference_table <- data.table::fread(paste0(data_dir, "reference table.csv"));
prepare_symbol_table <- function(sym, ref) {
# This check is only necessary if calling individually
if(data.table::uniqueN(ref$symbol) > 1)
ref <- ref[symbol == sym]
symbol_csv <- data.table::fread(paste0(data_dir, sym, ".csv"))
data.table::merge.data.table(ref, symbol_csv, by.x = c("showdate"), by.y = c("date"))
}
# merge a single symbol table
yum_table <- prepare_symbol_table("YUM", reference_table)
# all merged at once, reading individual CSVs by matching the symbol column from
# the reference table
all_symbols_merged <- reference_table[, {
# symbol_csv <- data.table::fread(paste0(data_dir, symbol, ".csv"))
# data.table::merge.data.table(.SD, symbol_csv, by.x = c("showdate"), by.y = c("date"))
prepare_symbol_table(.BY, .SD)
}, by = c("symbol")]