I am struggling to understand why legend.horizontal is not rotating my legend axis so it isn't displaying vertically? Any help would be massively appreciated.
library(phyloseq)
library(ggplot2)
##phylum level
ps_tmp <- get_top_taxa(physeq_obj = ps.phyl, n = 10, relative = TRUE, discard_other = FALSE, other_label = "Other")
ps_tmp <- name_taxa(ps_tmp, label = "Unkown", species = T, other_label = "Other")
phyl <- fantaxtic_bar(ps_tmp, color_by = "phylum", label_by = "phylum",facet_by = "TREATMENT", other_label = "Other", order_alg = "as.is")
phyl theme(legend.direction = "horizontal", legend.position = "bottom", )
CodePudding user response:
Legends for discrete values don't have a formal direction per se and are positioned however ggplot2 decides it can best fit with your data. This is why things like legend.direction won't work here. I don't have the phyloseq package or access to your particular data, so I'll show you how this works and how you can mess with the legend using a reproducible example dataset.
library(ggplot2)
set.seed(8675309)
df <- data.frame(x=LETTERS[1:8], y=sample(1:100, 8))
p <- ggplot(df, aes(x, y, fill=x)) geom_col()
p
By default, ggplot is putting our legend to the right and organizes it vertically as one column. Here's what happens when we move the legend to the bottom:
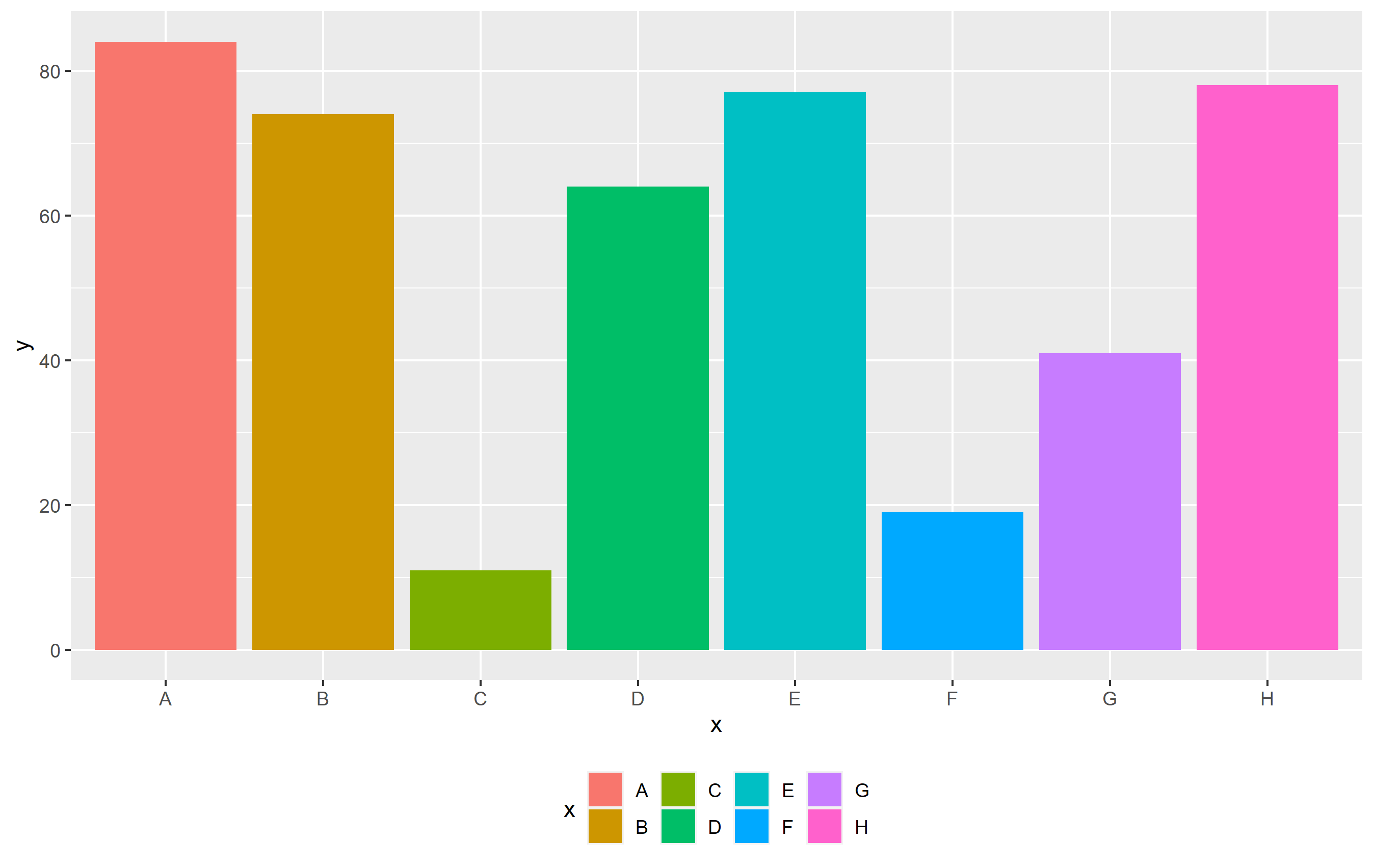
p theme(legend.position="bottom")
Now ggplot thinks it's best to put that legend into 4 columns, 2 rows each. As u/Tech Commodities mentioned, you can use the guides() functions to specify how the legend looks. In this case, we will specify to have 2 columns instead of 4. We only need to supply the number of columns (or rows), and ggplot figures out the rest.
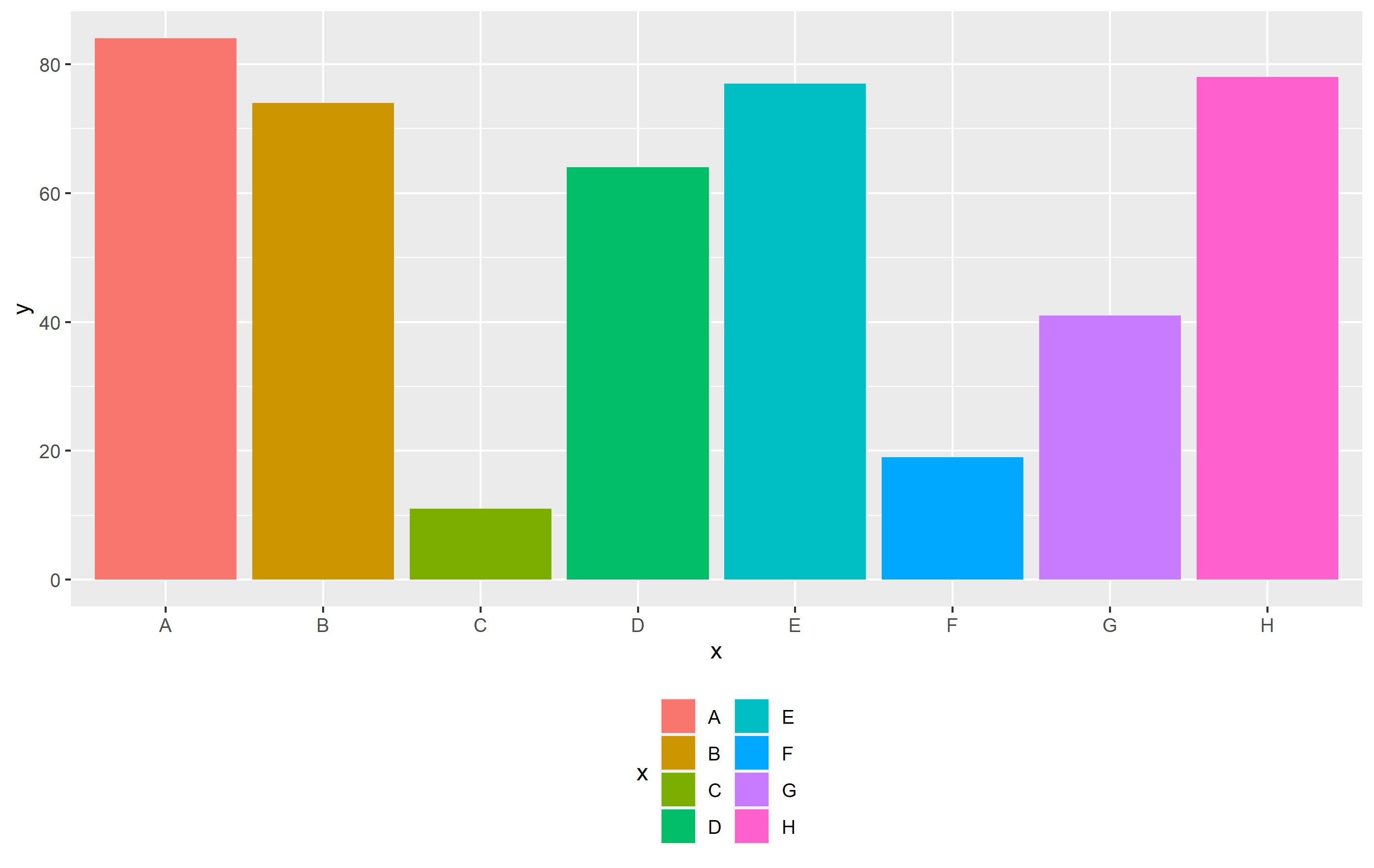
p theme(legend.position="bottom")
guides(fill=guide_legend(ncol=2))
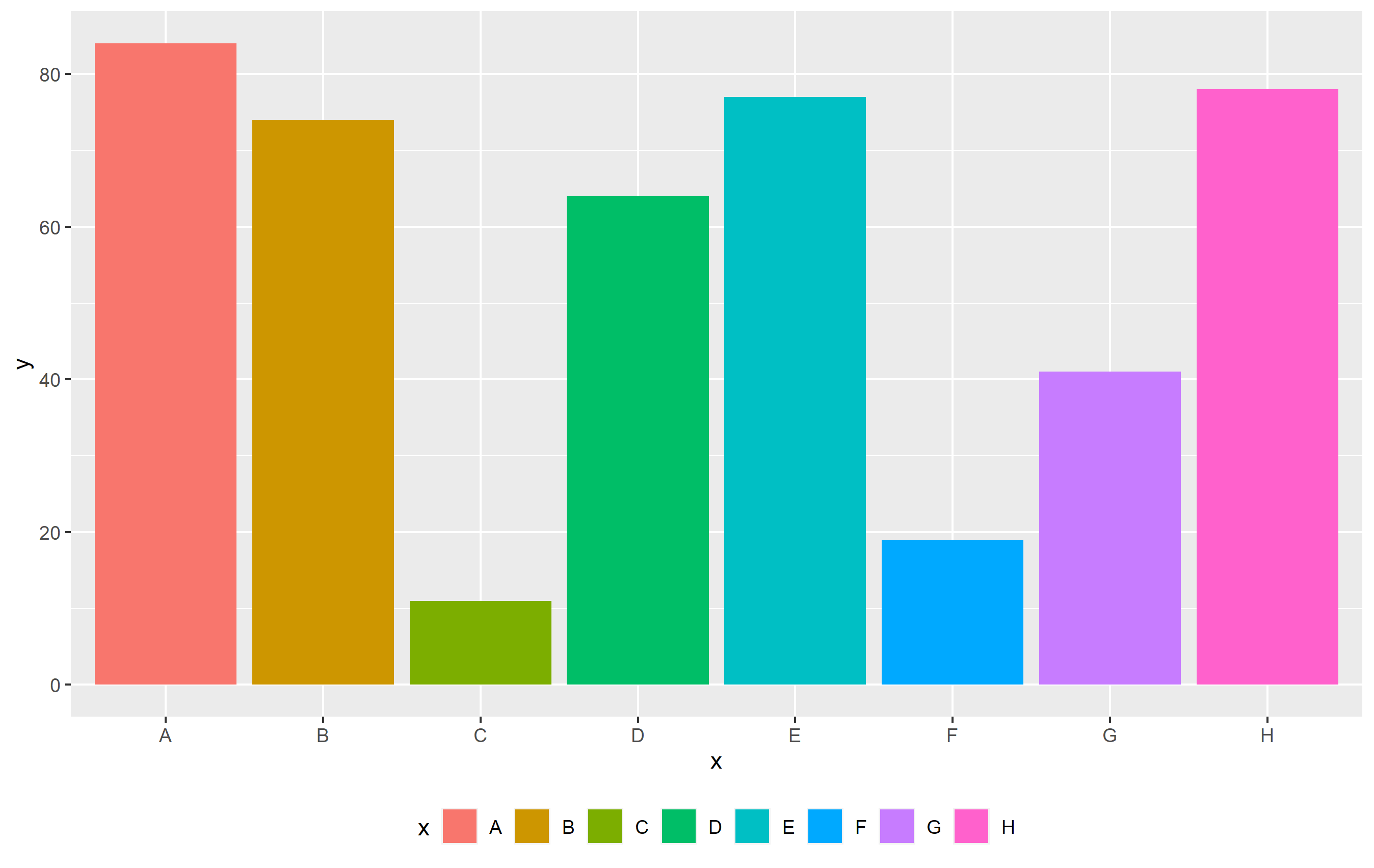
So, to get a "horizontally-arranged" legend, you just need to specify that there should be only one row:
p theme(legend.position="bottom")
guides(fill=guide_legend(nrow=1))